3UH8
 
 | | N-terminal domain of phage TP901-1 ORF48 | | Descriptor: | ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V.I, van Sinderen, D, Cambillau, C. | | Deposit date: | 2011-11-03 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1E5P
 
 | | Crystal structure of aphrodisin, a sex pheromone from female hamster | | Descriptor: | APHRODISIN | | Authors: | Vincent, F, Brown, K, Spinelli, S, Cambillau, C, Tegoni, M. | | Deposit date: | 2000-07-28 | | Release date: | 2001-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of aphrodisin, a sex pheromone from female hamster.
J.Mol.Biol., 305, 2001
|
|
4JEI
 
 | | Nonglycosylated Yarrowia lipolytica LIP2 lipase | | Descriptor: | Lipase 2 | | Authors: | Aloulou, A, Benarouche, A, Puccinelli, D, Spinelli, S, Cavalier, J.-F, Cambillau, C, Carriere, F. | | Deposit date: | 2013-02-27 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and structural characterization of non-glycosylated Yarrowia lipolytica LIP2 lipase
To be published
|
|
4HH6
 
 | | Peptide from EAEC T6SS Sci1 SciI protein | | Descriptor: | Peptide from EAEC T6SS Sci1 SciI protein, Putative type VI secretion protein | | Authors: | Douzi, B, Spinelli, S, Legrand, P, Lensi, V, Brunet, Y.R, Cascales, E, Cambillau, C. | | Deposit date: | 2012-10-09 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Role and specificity of ClpV ATPases in T6SS secretion.
To be Published
|
|
4GQK
 
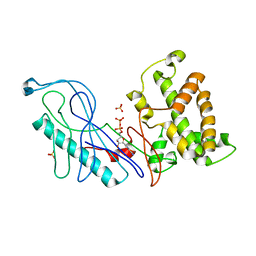 | | Structure of Native VgrG1-ACD with ADP (no cations) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, VgrG protein | | Authors: | Nguyen, V.S, Spinelli, S, Derrez, E, Durand, E, Cambillau, C. | | Deposit date: | 2012-08-23 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure of Native VgrG1-ACD with ADP (no cations)
To be Published
|
|
4GRW
 
 | | Structure of a complex of human IL-23 with 3 Nanobodies (Llama vHHs) | | Descriptor: | Interleukin-12 subunit beta, Interleukin-23 subunit alpha, Nanobody 124C4, ... | | Authors: | Desmyter, A, Spinelli, S, Button, C, Saunders, M, de Haard, H, Rommelaere, H, Union, A, Cambillau, C. | | Deposit date: | 2012-08-27 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Neutralization of Human Interleukin 23 by Multivalent Nanobodies Explained by the Structure of Cytokine-Nanobody Complex.
Front Immunol, 8, 2017
|
|
4HH5
 
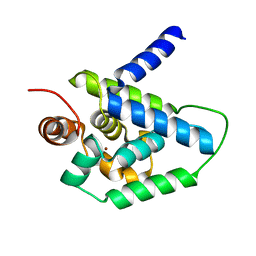 | | N-terminal domain (1-163) of ClpV1 ATPase from E.coli EAEC Sci1 T6SS. | | Descriptor: | BROMIDE ION, Putative type VI secretion protein | | Authors: | Douzi, B, Spinelli, S, Legrand, P, Lensi, V, Brunet, Y.R, Cascales, E, Cambillau, C. | | Deposit date: | 2012-10-09 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role and specificity of ClpV ATPases in T6SS secretion.
To be Published
|
|
1C12
 
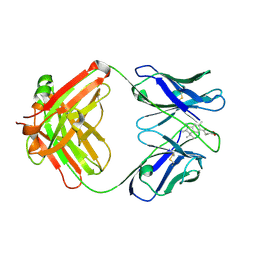 | | INSIGHT IN ODORANT PERCEPTION: THE CRYSTAL STRUCTURE AND BINDING CHARACTERISTICS OF ANTIBODY FRAGMENTS DIRECTED AGAINST THE MUSK ODORANT TRASEOLIDE | | Descriptor: | PROTEIN (ANTIBODY FRAGMENT FAB), TRAZEOLIDE | | Authors: | Langedijk, A.C, Spinelli, S, Anguille, C, Hermans, P, Nederlof, J, Butenandt, J, Honegger, A, Cambillau, C, Pluckthun, A. | | Deposit date: | 1999-07-20 | | Release date: | 1999-08-14 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into odorant perception: the crystal structure and binding characteristics of antibody fragments directed against the musk odorant traseolide.
J.Mol.Biol., 292, 1999
|
|
4E1F
 
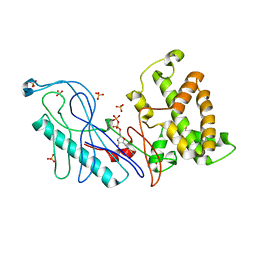 | | Structure of a VgrG Vibrio cholerae toxin ACD domain Glu16Gln mutant in complex with ADP and Mn++ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Durand, E, Audoly, G, Derrez, E, Spinelli, S, Ortiz-Lombardia, M, Cascales, E, Raoult, D, Cambillau, C. | | Deposit date: | 2012-03-06 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and functional characterization of the Vibrio cholerae toxin
from the VgrG/MARTX family.
J.Biol.Chem., 2012
|
|
4DTD
 
 | | Structure and functional characterization of a Vibrio cholerae toxin from the MARTX/VgrG family. | | Descriptor: | GLYCEROL, VgrG protein | | Authors: | Durand, E, Audoly, G, Derrez, E, Spinelli, S, Ortiz-Lombardia, M, Cascales, E, Raoult, D, Cambillau, C. | | Deposit date: | 2012-02-21 | | Release date: | 2012-08-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and functional characterization of a Vibrio cholerae toxin
from the MARTX/VgrG family.
J.Biol.Chem., 2012
|
|
4DTL
 
 | | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with ATP and Mn++ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Durand, E, Audoly, G, Derrez, E, Spinelli, S, Ortiz-Lombardia, M, Cascales, E, Raoult, D, Cambillau, C. | | Deposit date: | 2012-02-21 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with ATP and Mn++
J.Biol.Chem., 2012
|
|
4DM5
 
 | |
4DTH
 
 | | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with ATP and Mg++ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Durand, E, Audoly, G, Derrez, E, Spinelli, S, Ortiz-Lombardia, M, Cascales, E, Raoult, D, Cambillau, C. | | Deposit date: | 2012-02-21 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with ATP and Mg++
J.Biol.Chem., 2012
|
|
4E1D
 
 | | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with ADP and Mn++ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Durand, E, Audoly, G, Derrez, E, Spinelli, S, Ortiz-Lombardia, M, Cascales, E, Raoult, D, Cambillau, C. | | Deposit date: | 2012-03-06 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure and functional characterization of the Vibrio cholerae toxin
from the VgrG/MARTX family.
J.Biol.Chem., 2012
|
|
4DTF
 
 | | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with AMP-PNP and Mg++ | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Durand, E, Audoly, G, Derrez, E, Spinelli, S, Ortiz-Lombardia, M, Cascales, E, Raoult, D, Cambillau, C. | | Deposit date: | 2012-02-21 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with AMP-PNP and Mg++
J.Biol.Chem., 2012
|
|
4E1C
 
 | | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with ADP and Mg++ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Durand, E, Audoly, G, Derrez, E, Spinelli, S, Ortiz-Lombardia, M, Cascales, E, Raoult, D, Cambillau, C. | | Deposit date: | 2012-03-06 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and functional characterization of the Vibrio cholerae toxin
from the VgrG/MARTX family.
J.Biol.Chem., 2012
|
|
3R1V
 
 | | Odorant Binding Protein 7 from Anopheles gambiae with Four Disulfide Bridges, in complex with an azo compound | | Descriptor: | 4-{(E)-[4-(propan-2-yl)phenyl]diazenyl}phenol, Odorant binding protein, antennal | | Authors: | Lagarde, A, Spinelli, S, Tegoni, M, Field, L, He, X, Zhou, J.J, Cambillau, C. | | Deposit date: | 2011-03-11 | | Release date: | 2011-10-19 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Crystal Structure of Odorant Binding Protein 7 from Anopheles gambiae Exhibits an Outstanding Adaptability of Its Binding Site.
J.Mol.Biol., 414, 2011
|
|
3R1O
 
 | | Odorant Binding Protein 7 from Anopheles gambiae with Four Disulfide Bridges | | Descriptor: | Odorant binding protein, antennal, PALMITIC ACID | | Authors: | Lagarde, A, Spinelli, S, Tegoni, M, Field, L, He, X, Zhou, J.J, Cambillau, C. | | Deposit date: | 2011-03-11 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Odorant Binding Protein 7 from Anopheles gambiae Exhibits an Outstanding Adaptability of Its Binding Site.
J.Mol.Biol., 414, 2011
|
|
3R1P
 
 | | Odorant Binding Protein 7 from Anopheles gambiae with Four Disulfide Bridges, form P1 | | Descriptor: | Odorant binding protein, antennal, PALMITIC ACID | | Authors: | Lagarde, A, Spinelli, S, Tegoni, M, Field, L, He, X, Zhou, J.J, Cambillau, C. | | Deposit date: | 2011-03-11 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of Odorant Binding Protein 7 from Anopheles gambiae Exhibits an Outstanding Adaptability of Its Binding Site.
J.Mol.Biol., 414, 2011
|
|
3U66
 
 | | Crystal structure of T6SS SciP/TssL from Escherichia Coli Enteroaggregative 042 | | Descriptor: | GLYCEROL, Putative type VI secretion protein | | Authors: | Durand, E, Aschtgen, M.S, Zoued, A, Spinelli, S, Watson, P.J.H, Cambillau, C, Cascales, E. | | Deposit date: | 2011-10-12 | | Release date: | 2012-03-07 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural characterization and oligomerization of the TssL protein, a component shared by bacterial type VI and type IVb secretion systems.
J.Biol.Chem., 287, 2012
|
|
2YEY
 
 | | Crystal structure of the allosteric-defective chaperonin GroEL E434K mutant | | Descriptor: | 60 KDA CHAPERONIN | | Authors: | Cabo-Bilbao, A, Mechaly, A.E, Agirre, J, Spinelli, S, Sot, B, Muga, A, Guerin, D.M.A. | | Deposit date: | 2011-03-31 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Crystal Structure of the Temperature-Sensitive and Allosteric-Defective Chaperonin Groele461K.
J.Struct.Biol., 155, 2006
|
|
3BFB
 
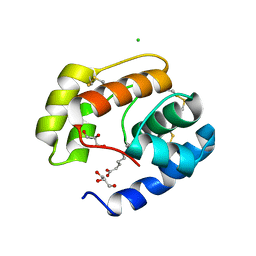 | | Crystal structure of a pheromone binding protein from Apis mellifera in complex with the 9-keto-2(E)-decenoic acid | | Descriptor: | (2Z)-9-oxodec-2-enoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2007-11-21 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
3BFH
 
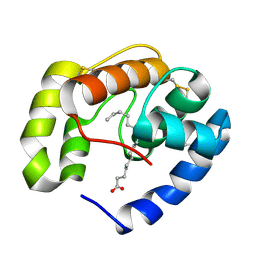 | | Crystal structure of a pheromone binding protein from Apis mellifera in complex with hexadecanoic acid | | Descriptor: | CHLORIDE ION, PALMITIC ACID, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2007-11-21 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
3BFA
 
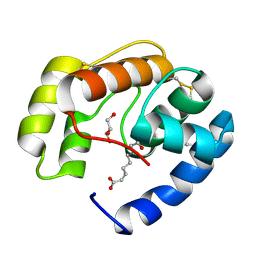 | | Crystal structure of a pheromone binding protein from Apis mellifera in complex with the Queen mandibular pheromone | | Descriptor: | (2Z)-9-oxodec-2-enoic acid, GLYCEROL, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2007-11-21 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
3CAB
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera soaked at pH 7.0 | | Descriptor: | GLYCEROL, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-02-19 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
