6DMZ
 
 | | Solution structure of ZmD32 | | Descriptor: | Flower-specific gamma-thionin | | Authors: | Harvey, P.J, Craik, D.J. | | Deposit date: | 2018-06-05 | | Release date: | 2019-05-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Salt-Tolerant Antifungal and Antibacterial Activities of the Corn Defensin ZmD32.
Front Microbiol, 10, 2019
|
|
2JYY
 
 | |
2J15
 
 | | Cyclic MrIA: An exceptionally stable and potent cyclic conotoxin with a novel topological fold that targets the norepinephrine transporter. | | Descriptor: | MAI126P | | Authors: | Lovelace, E.S, Armishaw, C.J, Colgrave, M.L, Walstrom, M.E, Alewood, P.F, Daly, N.L, Craik, D.J. | | Deposit date: | 2006-08-09 | | Release date: | 2006-11-01 | | Last modified: | 2018-05-09 | | Method: | SOLUTION NMR | | Cite: | Cyclic MrIA: a stable and potent cyclic conotoxin with a novel topological fold that targets the norepinephrine transporter.
J. Med. Chem., 49, 2006
|
|
2KHB
 
 | |
2KRA
 
 | |
2GJ0
 
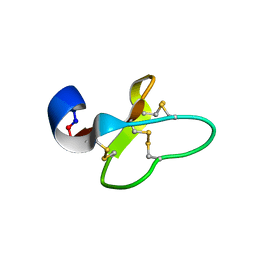 | | Cycloviolacin O14 | | Descriptor: | Cycloviolacin O14 | | Authors: | Ireland, D.C, Colgrave, M.L, Craik, D.J. | | Deposit date: | 2006-03-30 | | Release date: | 2006-04-11 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | A novel suite of cyclotides from Viola odorata: sequence variation and the implications for structure, function and stability
Biochem.J., 400, 2006
|
|
2GW9
 
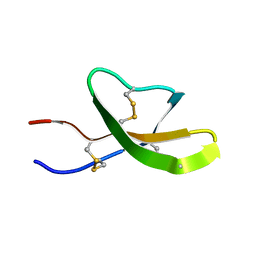 | | High-resolution solution structure of the mouse defensin Cryptdin4 | | Descriptor: | Defensin-related cryptdin 4 | | Authors: | Rosengren, K.J, Craik, D.J, Vogel, H.J, Daly, N.L, Ouellette, A.J. | | Deposit date: | 2006-05-04 | | Release date: | 2006-07-25 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of the conserved salt bridge in mammalian paneth cell alpha-defensins: solution structures of mouse CRYPTDIN-4 and (E15D)-CRYPTDIN-4.
J.Biol.Chem., 281, 2006
|
|
2GWP
 
 | | High-resolution solution structure of the salt-bridge defficient mouse defensin (E15D)-Cryptdin4 | | Descriptor: | Defensin-related cryptdin 4 | | Authors: | Rosengren, K.J, Craik, D.J, Vogel, H.J, Daly, N.L, Ouellette, A.J. | | Deposit date: | 2006-05-05 | | Release date: | 2006-07-25 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of the conserved salt bridge in mammalian paneth cell alpha-defensins: solution structures of mouse CRYPTDIN-4 and (E15D)-CRYPTDIN-4.
J.Biol.Chem., 281, 2006
|
|
2KUK
 
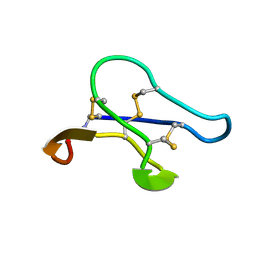 | |
2KUX
 
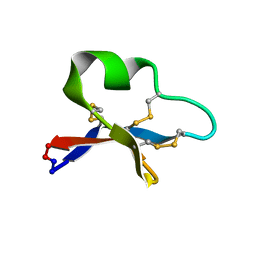 | |
2LR5
 
 | |
2LJS
 
 | | Solution structure of MCoTI-V | | Descriptor: | Trypsin inhibitor 3 | | Authors: | Daly, N.L, Craik, D.J. | | Deposit date: | 2011-09-23 | | Release date: | 2012-08-08 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Cyclic Peptides Arising by Evolutionary Parallelism via Asparaginyl-Endopeptidase-Mediated Biosynthesis.
Plant Cell, 24, 2012
|
|
2LEY
 
 | | Solution structure of (R7G)-Crp4 | | Descriptor: | Alpha-defensin 4 | | Authors: | Rosengren, K, Andersson, H.S, Haugaard-Kedstrom, L.M, Bengtsson, E, Daly, N.L, Figueredo, S.M, Qu, X, Craik, D.J, Ouellette, A.J. | | Deposit date: | 2011-06-26 | | Release date: | 2012-05-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The alpha-defensin salt-bridge induces backbone stability to facilitate folding and confer proteolytic resistance.
Amino Acids, 43, 2012
|
|
2LEW
 
 | | Structural Plasticity of Paneth cell alpha-Defensins: Characterization of Salt-Bridge Deficient Analogues of Mouse Cryptdin-4 | | Descriptor: | Alpha-defensin 4 | | Authors: | Rosengren, K, Andersson, H.S, Haugaard-Kedstrom, L.M, Bengtsson, E, Daly, N.L, Craik, D.J. | | Deposit date: | 2011-06-24 | | Release date: | 2012-05-16 | | Last modified: | 2013-06-19 | | Method: | SOLUTION NMR | | Cite: | The alpha-defensin salt-bridge induces backbone stability to facilitate folding and confer proteolytic resistance.
Amino Acids, 43, 2012
|
|
2MSQ
 
 | | Solution study of cBru9a | | Descriptor: | Conotoxin cBru9a | | Authors: | Akcan, M, Clark, R.J, Daly, N.L, Conibear, A.C, de Faoite, A.C, Heghinian, M.C, Adams, D.J, Mari, F, Craik, D.J. | | Deposit date: | 2014-08-05 | | Release date: | 2015-07-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Transforming conotoxins into cyclotides: Backbone cyclization of P-superfamily conotoxins.
Biopolymers, 104, 2015
|
|
2NS3
 
 | | Solution structure of ribbon BuIA | | Descriptor: | Alpha-conotoxin BuIA | | Authors: | Jin, A.H, Brandstaetter, H, Nevin, S.T, Tan, C.C, Clark, R.J, Adams, D.J, Alewood, P.F, Craik, D.J, Daly, N.L. | | Deposit date: | 2006-11-03 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of alpha-conotoxin BuIA: influences of disulfide connectivity on structural dynamics
Bmc Struct.Biol., 7, 2007
|
|
2MSO
 
 | | Solution study of cGm9a | | Descriptor: | Conotoxin Gm9.1 | | Authors: | Akcan, M, Clark, R.J, Daly, N.L, Conibear, A.C, de Faoite, A.C, Heghinian, M.C, Adams, D.J, Mari, F, Craik, D.J. | | Deposit date: | 2014-08-05 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Transforming conotoxins into cyclotides: Backbone cyclization of P-superfamily conotoxins.
Biopolymers, 104, 2015
|
|
2NAW
 
 | |
2NAV
 
 | | NMR solution structure of Ex-4[1-16]/pl14a | | Descriptor: | Exendin-4, Alpha/kappa-conotoxin pl14a chimera | | Authors: | Schroeder, C.I, Swedberg, J.E, Craik, D.J. | | Deposit date: | 2016-01-11 | | Release date: | 2016-05-04 | | Last modified: | 2016-08-17 | | Method: | SOLUTION NMR | | Cite: | Truncated Glucagon-like Peptide-1 and Exendin-4 alpha-Conotoxin pl14a Peptide Chimeras Maintain Potency and alpha-Helicity and Reveal Interactions Vital for cAMP Signaling in Vitro.
J.Biol.Chem., 291, 2016
|
|
1AG7
 
 | | CONOTOXIN GS, NMR, 20 STRUCTURES | | Descriptor: | CONOTOXIN GS | | Authors: | Hill, J.M, Alewood, P.F, Craik, D.J. | | Deposit date: | 1997-04-03 | | Release date: | 1998-04-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the sodium channel antagonist conotoxin GS: a new molecular caliper for probing sodium channel geometry.
Structure, 5, 1997
|
|
1BH4
 
 | |
1BJC
 
 | | SOLUTION NMR STRUCTURE OF AMYLOID BETA[F16], RESIDUES 1-28, 15 STRUCTURES | | Descriptor: | AMYLOID BETA-PEPTIDE | | Authors: | Poulsen, S.-A, Watson, A.A, Craik, D.J. | | Deposit date: | 1998-06-23 | | Release date: | 1998-11-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures in aqueous SDS micelles of two amyloid beta peptides of A beta(1-28) mutated at the alpha-secretase cleavage site (K16E, K16F)
J.Struct.Biol., 130, 2000
|
|
1CE3
 
 | |
1BA4
 
 | | THE SOLUTION STRUCTURE OF AMYLOID BETA-PEPTIDE (1-40) IN A WATER-MICELLE ENVIRONMENT. IS THE MEMBRANE-SPANNING DOMAIN WHERE WE THINK IT IS? NMR, 10 STRUCTURES | | Descriptor: | AMYLOID BETA-PEPTIDE | | Authors: | Coles, M, Bicknell, W, Watson, A.A, Fairlie, D.P, Craik, D.J. | | Deposit date: | 1998-04-07 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of amyloid beta-peptide(1-40) in a water-micelle environment. Is the membrane-spanning domain where we think it is?
Biochemistry, 37, 1998
|
|
1CNN
 
 | | OMEGA-CONOTOXIN MVIIC FROM CONUS MAGUS | | Descriptor: | OMEGA-CONOTOXIN MVIIC | | Authors: | Nielsen, K.J, Adams, D, Thomas, L, Bond, T, Alewood, P.F, Craik, D.J, Lewis, R.J. | | Deposit date: | 1999-05-20 | | Release date: | 2000-05-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure-activity relationships of omega-conotoxins MVIIA, MVIIC and 14 loop splice hybrids at N and P/Q-type calcium channels.
J.Mol.Biol., 289, 1999
|
|
