3A55
 
 | |
1CSC
 
 | |
1CSI
 
 | |
1D1P
 
 | | CRYSTAL STRUCTURE OF A YEAST LOW MOLECULAR WEIGHT PROTEIN TYROSINE PHOSPHATASE (LTP1) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TYROSINE PHOSPHATASE | | Authors: | Wang, S, Tabernero, L, Zhang, M, Harms, E, Van Etten, R.L, Stauffacher, C.V. | | Deposit date: | 1999-09-20 | | Release date: | 2000-03-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of a low-molecular weight protein tyrosine phosphatase from Saccharomyces cerevisiae and its complex with the substrate p-nitrophenyl phosphate.
Biochemistry, 39, 2000
|
|
1D1Q
 
 | | CRYSTAL STRUCTURE OF A YEAST LOW MOLECULAR WEIGHT PROTEIN TYROSINE PHOSPHATASE (LTP1) COMPLEXED WITH THE SUBSTRATE PNPP | | Descriptor: | 4-NITROPHENYL PHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Wang, S, Tabernero, L, Zhang, M, Harms, E, Van Etten, R.L, Staufacher, C.V. | | Deposit date: | 1999-09-20 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of a low-molecular weight protein tyrosine phosphatase from Saccharomyces cerevisiae and its complex with the substrate p-nitrophenyl phosphate.
Biochemistry, 39, 2000
|
|
1D2A
 
 | | CRYSTAL STRUCTURE OF A YEAST LOW MOLECULAR WEIGHT PROTEIN TYROSINE PHOSPHATASE (LTP1) COMPLEXED WITH THE ACTIVATOR ADENINE | | Descriptor: | ADENINE, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Wang, S, Stauffacher, C.V, Van Etten, R.L. | | Deposit date: | 1999-09-22 | | Release date: | 2000-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic basis for the activation of a low-molecular weight protein tyrosine phosphatase by adenine.
Biochemistry, 39, 2000
|
|
2ZL8
 
 | |
1CSH
 
 | |
1G4A
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
1G4B
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | Descriptor: | ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
2ZWM
 
 | |
2ZXJ
 
 | |
1VC4
 
 | | Crystal Structure of Indole-3-Glycerol Phosphate Synthase (TrpC) from Thermus Thermophilus At 1.8 A Resolution | | Descriptor: | ACETIC ACID, GLYCEROL, Indole-3-Glycerol Phosphate Synthase, ... | | Authors: | Bagautdinov, B, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-04 | | Release date: | 2004-03-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of indole-3-glycerol phosphate synthase from Thermus thermophilus HB8: implications for thermal stability.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1CTS
 
 | |
3AHN
 
 | | PZ PEPTIDASE A with Inhibitor 1 | | Descriptor: | ACETATE ION, N~2~-{(2S)-3-[(R)-hydroxy{(1R)-2-phenyl-1-[(phenylacetyl)amino]ethyl}phosphoryl]-2-methylpropanoyl}-L-lysyl-D-serine, Oligopeptidase, ... | | Authors: | Nakano, H. | | Deposit date: | 2010-04-25 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The exquisite structure and reaction mechanism of bacterial Pz-peptidase A toward collagenous peptides: X-ray crystallographic structure analysis of PZ-peptidase a reveals differences from mammalian thimet oligopeptidase.
J.Biol.Chem., 285, 2010
|
|
6M6V
 
 | | Crystal structure the toxin-antitoxin MntA-HepT | | Descriptor: | RNA (5'-R(P*AP*AP*A)-3'), Toxin-antitoxin system antidote Mnt family, Toxin-antitoxin system toxin HepN family | | Authors: | Ouyang, S.Y, Zhen, X.K. | | Deposit date: | 2020-03-16 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Novel polyadenylylation-dependent neutralization mechanism of the HEPN/MNT toxin/antitoxin system.
Nucleic Acids Res., 48, 2020
|
|
6M6W
 
 | | Crystal structure the toxin-antitoxin MntA-HpeT mutant-Y104A | | Descriptor: | Toxin-antitoxin system antidote Mnt family, Toxin-antitoxin system toxin HepN family | | Authors: | Ouyang, S.Y, Zhen, X.K. | | Deposit date: | 2020-03-16 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.611 Å) | | Cite: | Novel polyadenylylation-dependent neutralization mechanism of the HEPN/MNT toxin/antitoxin system.
Nucleic Acids Res., 48, 2020
|
|
6M6U
 
 | | Crystal structure the toxin-antitoxin MntA-HpeT mutant-D39ED41E | | Descriptor: | Toxin-antitoxin system antitoxin MntA family, Toxin-antitoxin system toxin HepN family | | Authors: | Ouyang, S.Y, Zhen, X.K. | | Deposit date: | 2020-03-16 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Novel polyadenylylation-dependent neutralization mechanism of the HEPN/MNT toxin/antitoxin system.
Nucleic Acids Res., 48, 2020
|
|
3AHO
 
 | | PZ PEPTIDASE A with inhibitor 2 | | Descriptor: | 1-{3-[(R)-{(1R)-1-[(glycyl-L-prolyl)amino]-2-phenylethyl}(hydroxy)phosphoryl]propanoyl}-L-prolyl-D-norleucine, ACETATE ION, Oligopeptidase, ... | | Authors: | Nakano, H. | | Deposit date: | 2010-04-25 | | Release date: | 2010-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The exquisite structure and reaction mechanism of bacterial Pz-peptidase A toward collagenous peptides: X-ray crystallographic structure analysis of PZ-peptidase a reveals differences from mammalian thimet oligopeptidase.
J.Biol.Chem., 285, 2010
|
|
3WHX
 
 | | Crystal structure of anti-prostaglandin E2 Fab fragment PGE1 complex | | Descriptor: | 7-[(1R,3R)-3-hydroxy-2-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-5-oxocyclopentyl]heptanoic acid, mAb Fab H fragment, mAb Fab L fragment | | Authors: | Sugahara, M, Ago, H, Saino, H, Miyano, M. | | Deposit date: | 2013-09-03 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of anti-Prostaglandin E2 Fab fragment with Prostaglandin E2
To be Published
|
|
3WIF
 
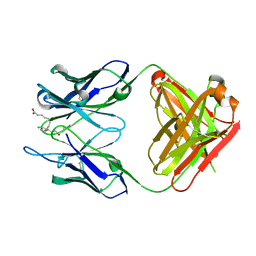 | | Crystal structure of anti-prostaglandin E2 Fab fragment 9Cl-PGF2beta complex | | Descriptor: | (Z)-7-[(1R,2R,3R,5R)-5-chloranyl-3-oxidanyl-2-[(E,3S)-3-oxidanyloct-1-enyl]cyclopentyl]hept-5-enoic acid, mAb Fab H fragment, mAb Fab L fragment | | Authors: | Sugahara, M, Ago, H, Saino, H, Miyano, M. | | Deposit date: | 2013-09-12 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of anti-Prostaglandin E2 Fab fragment with Prostaglandin E2
To be Published
|
|
3WE6
 
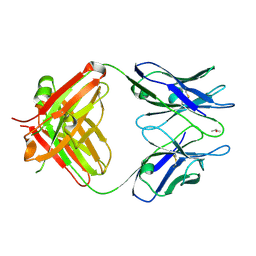 | |
3WFH
 
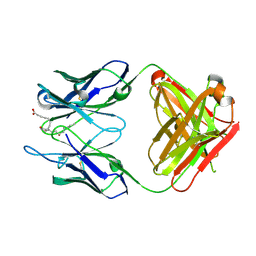 | | Crystal structure of anti-Prostaglandin E2 Fab fragment PGE2 complex | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, mAb Fab H fragment, mAb Fab L fragment | | Authors: | Sugahara, M, Ago, H, Saino, H, Miyano, M. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of anti-Prostaglandin E2 Fab fragment with Prostaglandin E2
To be Published
|
|
3BYV
 
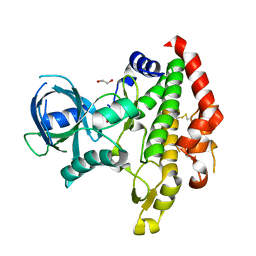 | | Crystal structure of Toxoplasma gondii specific rhoptry antigen kinase domain | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Rhoptry kinase | | Authors: | Wernimont, A.K, Lunin, V.V, Yang, C, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Sibley, D, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-16 | | Release date: | 2008-01-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel structural and regulatory features of rhoptry secretory kinases in Toxoplasma gondii.
Embo J., 28, 2009
|
|
6LF1
 
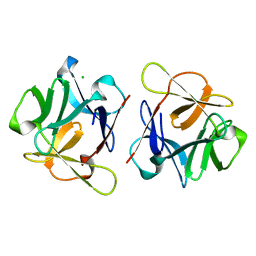 | | SeviL, a GM1b/asialo-GM1 binding lectin | | Descriptor: | CHLORIDE ION, SeviL | | Authors: | Kamata, K, Ozeki, Y, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2019-11-28 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of SeviL, a GM1b/asialo-GM1 binding R-type lectin from the mussel Mytilisepta virgata.
Sci Rep, 10, 2020
|
|
