6UGB
 
 | | C3 symmetric peptide design number 2, Baby Basil | | Descriptor: | C3 symmetric peptide design number 2, Baby Basil, CHLORIDE ION, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
5EB9
 
 | | Crystal Structure Of Chicken CD8aa Homodimer | | Descriptor: | CD8 alpha chain | | Authors: | Liu, Y.J, Qi, J.X, Xia, C. | | Deposit date: | 2015-10-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | The structural basis of chicken, swine and bovine CD8 alpha alpha dimers provides insight into the co-evolution with MHC I in endotherm species.
Sci Rep, 6, 2016
|
|
3HW4
 
 | | Crystal structure of avian influenza A virus in complex with TMP | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, THYMIDINE-5'-PHOSPHATE | | Authors: | Zhao, C, Lou, Z, Guo, Y, Ma, M, Chen, Y, Rao, Z. | | Deposit date: | 2009-06-17 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleoside monophosphate complex structures of the endonuclease domain from the influenza virus polymerase PA subunit reveal the substrate binding site inside the catalytic center
J.Virol., 83, 2009
|
|
7CKF
 
 | | The N-terminus of interferon-inducible antiviral protein-dimer | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, Guanylate-binding protein 5, ... | | Authors: | Cui, W, Yang, H.T. | | Deposit date: | 2020-07-16 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.284 Å) | | Cite: | Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3HW5
 
 | | crystal structure of avian influenza virus PA_N in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Polymerase acidic protein | | Authors: | Zhao, C, Lou, Z, Guo, Y, Ma, M, Chen, Y, Liang, S, Rao, Z. | | Deposit date: | 2009-06-17 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Nucleoside monophosphate complex structures of the endonuclease domain from the influenza virus polymerase PA subunit reveal the substrate binding site inside the catalytic center
J.Virol., 83, 2009
|
|
6UF7
 
 | | S2 symmetric peptide design number 5, Uncle Fester | | Descriptor: | S2-5, Uncle Fester | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UF8
 
 | | S2 symmetric peptide design number 6, London Bridge | | Descriptor: | S2-6, London Bridge | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UDR
 
 | | S2 symmetric peptide design number 3 crystal form 1, Lurch | | Descriptor: | S2-3, Lurch crystal form 1 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UFU
 
 | | C2 symmetric peptide design number 1, Zappy, crystal form 1 | | Descriptor: | C2-1, Zappy, crystal form 1 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UG3
 
 | | C3 symmetric peptide design number 1, Sporty, crystal form 1 | | Descriptor: | C3-1, Sporty, crystal form 1, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UGC
 
 | | C3 symmetric peptide design number 3 | | Descriptor: | C3-3 cyclic peptide design, CADMIUM ION, SODIUM ION | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
5E00
 
 | | Structure of HLA-A2 P130 | | Descriptor: | Beta-2-microglobulin, GLY-VAL-TRP-ILE-ARG-THR-PRO-PRO-ALA, HLA class I histocompatibility antigen, ... | | Authors: | Zhang, Y, Wu, Y, Qi, J, Liu, J, Gao, G.F, Meng, S. | | Deposit date: | 2015-09-26 | | Release date: | 2017-01-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CD8+T-Cell Response-Associated Evolution of Hepatitis B Virus Core Protein and Disease Progress.
J. Virol., 92, 2018
|
|
3HW6
 
 | | Crystal structure of avian influenza virus PA_N in complex with Mn | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Zhao, C, Lou, Z, Guo, Y, Ma, M, Chen, Y, Rao, Z. | | Deposit date: | 2009-06-17 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nucleoside monophosphate complex structures of the endonuclease domain from the influenza virus polymerase PA subunit reveal the substrate binding site inside the catalytic center
J.Virol., 83, 2009
|
|
6DWZ
 
 | | Hermes transposase deletion dimer complex with (C/G) DNA | | Descriptor: | DNA (26-MER), DNA (5'-D(*GP*AP*GP*AP*AP*CP*AP*AP*CP*AP*AP*CP*AP*AP*G)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*C)-3'), ... | | Authors: | Dyda, F, Hickman, A.B. | | Deposit date: | 2018-06-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the mechanism of double strand break formation by Hermes, a hAT family eukaryotic DNA transposase.
Nucleic Acids Res., 46, 2018
|
|
8GSD
 
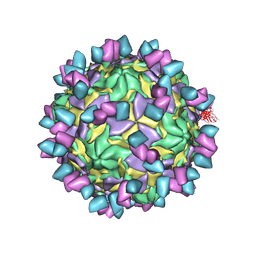 | | Echovirus3 full particle in complex with 6D10 Fab | | Descriptor: | Genome polyprotein, Genome polyprotein (Fragment), Heavy chain of 6D10, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-14 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis for the Immunogenicity of the C-Terminus of VP1 of Echovirus 3 Revealed by the Binding of a Neutralizing Antibody.
Viruses, 14, 2022
|
|
8GSF
 
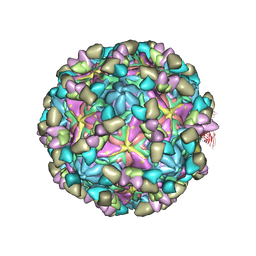 | | Echovirus3 empty particle in complex with 6D10 Fab (sideling) | | Descriptor: | Heavy chain of 6D10, Light chain of 6D10, VP0, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-14 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for the Immunogenicity of the C-Terminus of VP1 of Echovirus 3 Revealed by the Binding of a Neutralizing Antibody.
Viruses, 14, 2022
|
|
8GSE
 
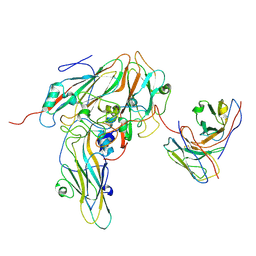 | | Echovirus3 capsid protein in complex with 6D10 Fab (upright) | | Descriptor: | Heavy chain of 6D10, Light chain of 6D10, VP0, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-14 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis for the Immunogenicity of the C-Terminus of VP1 of Echovirus 3 Revealed by the Binding of a Neutralizing Antibody.
Viruses, 14, 2022
|
|
8GSC
 
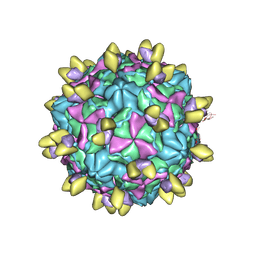 | | Echovirus3 A-particle in complex with 6D10 Fab | | Descriptor: | Heavy chain of 6D10, Light chain of 6D10, VP1, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-14 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis for the Immunogenicity of the C-Terminus of VP1 of Echovirus 3 Revealed by the Binding of a Neutralizing Antibody.
Viruses, 14, 2022
|
|
5Z65
 
 | | Crystal structure of porcine aminopeptidase N ectodomain in functional form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, Y.G, Li, R, Jiang, L.G, Qiao, S.L, Zhang, G.P. | | Deposit date: | 2018-01-22 | | Release date: | 2019-01-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Characterization of the interaction between recombinant porcine aminopeptidase N and spike glycoprotein of porcine epidemic diarrhea virus.
Int. J. Biol. Macromol., 117, 2018
|
|
8GU6
 
 | | Structure of the SbCas7-11-crRNA-NTR-Csx29 complex | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (33-MER), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-09-10 | | Release date: | 2023-01-18 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Target RNA-guided protease activity in type III-E CRISPR-Cas system.
Nucleic Acids Res., 50, 2022
|
|
8GNA
 
 | | Structure of the SbCas7-11-crRNA-NTR complex | | Descriptor: | RAMP superfamily protein, RNA (32-MER), RNA (5'-R(P*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*GP*GP*U)-3'), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-08-23 | | Release date: | 2023-01-18 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Target RNA-guided protease activity in type III-E CRISPR-Cas system.
Nucleic Acids Res., 50, 2022
|
|
6BUI
 
 | |
6BZ3
 
 | | Complex structure of FAK FAT domain and DCC P3 motif | | Descriptor: | CALCIUM ION, Focal adhesion kinase 1, Netrin receptor DCC | | Authors: | Xu, S, Wang, J.-H. | | Deposit date: | 2017-12-22 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | The binding of DCC-P3 motif and FAK-FAT domain mediates the initial step of netrin-1/DCC signaling for axon attraction.
Cell Discov, 4, 2018
|
|
6BTH
 
 | | Crystal structure of human cellular retinol binding protein 2 (CRBP2) in complex with 2-arachidonoylglycerol (2-AG) | | Descriptor: | 1,3-dihydroxypropan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, DI(HYDROXYETHYL)ETHER, Retinol-binding protein 2 | | Authors: | Silvaroli, J.A, Blaner, W.S, Lodowski, D.T, Golczak, M. | | Deposit date: | 2017-12-06 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Retinol-binding protein 2 (RBP2) binds monoacylglycerols and modulates gut endocrine signaling and body weight.
Sci Adv, 6, 2020
|
|
8GZ2
 
 | | Cryo-EM structure of human NaV1.6/beta1/beta2-4,9-anhydro-tetrodotoxin | | Descriptor: | (1R,2S,3S,4R,5R,9S,11S,12S,14R)-7-amino-2,4,12-trihydroxy-2-(hydroxymethyl)-10,13,15-trioxa-6,8-diazapentacyclo[7.4.1.1~3,12~.0~5,11~.0~5,14~]pentadec-7-en-8-ium (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein type 8 subunit alpha, ... | | Authors: | Li, Y, Jiang, D. | | Deposit date: | 2022-09-24 | | Release date: | 2023-03-08 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human Na V 1.6 channel reveals Na + selectivity and pore blockade by 4,9-anhydro-tetrodotoxin.
Nat Commun, 14, 2023
|
|
