6H5B
 
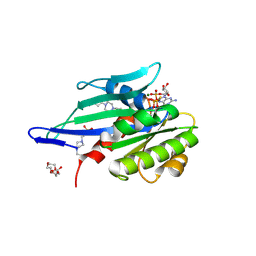 | | Myxococcus xanthus MglA in complex with its GAP MglB and GTPgammaS | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Galicia, C, Cherfils, J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-11-27 | | Last modified: | 2020-11-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | MglA functions as a three-state GTPase to control movement reversals of Myxococcus xanthus.
Nat Commun, 10, 2019
|
|
6ICW
 
 | |
6ID2
 
 | |
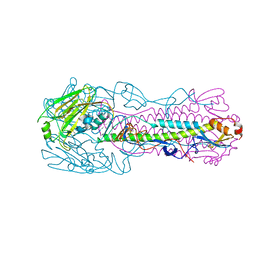
6K15
 
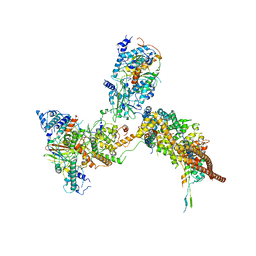 | | RSC substrate-recruitment module | | Descriptor: | Chromatin structure-remodeling complex protein RSC3, Chromatin structure-remodeling complex protein RSC30, Chromatin structure-remodeling complex protein RSC58, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-05-09 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome.
Science, 366, 2019
|
|
2XFB
 
 | | CHIKUNGUNYA E1 E2 ENVELOPE GLYCOPROTEINS FITTED IN SINDBIS VIRUS cryo- EM MAP | | Descriptor: | E1 ENVELOPE GLYCOPROTEIN, E2 ENVELOPE GLYCOPROTEIN | | Authors: | Voss, J.E, Vaney, M.C, Duquerroy, S, Rey, F.A. | | Deposit date: | 2010-05-21 | | Release date: | 2010-11-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Glycoprotein Organization of Chikungunya Virus Particles Revealed by X-Ray Crystallography
Nature, 468, 2010
|
|
3IY9
 
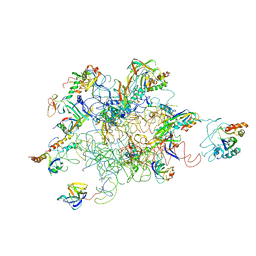 | | Leishmania Tarentolae Mitochondrial Large Ribosomal Subunit Model | | Descriptor: | 39S ribosomal protein L11, mitochondrial, 39S ribosomal protein L16, ... | | Authors: | Sharma, M.R, Booth, T.M, Simpson, L, Maslov, D.A, Agrawal, R.K. | | Deposit date: | 2009-04-20 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Structure of a mitochondrial ribosome with minimal RNA
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7VA8
 
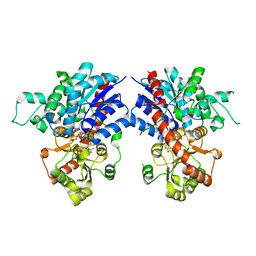 | | Crystal structure of MiCGT | | Descriptor: | UDP-glycosyltransferase 13, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Zhong, L, Zhang, Z.M. | | Deposit date: | 2021-08-27 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85003233 Å) | | Cite: | Directed Evolution of a Plant Glycosyltransferase for Chemo- and Regioselective Glycosylation of Pharmaceutically Significant Flavonoids
Acs Catalysis, 11, 2021
|
|
7VAA
 
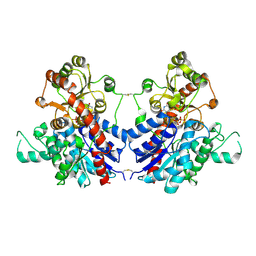 | |
4NL1
 
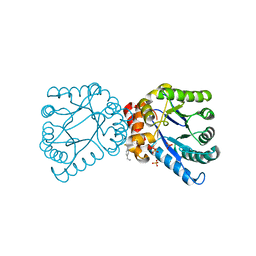 | |
4NHV
 
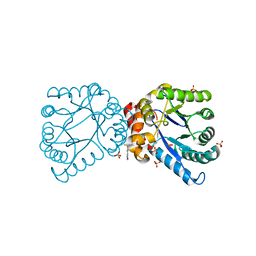 | |
4NIL
 
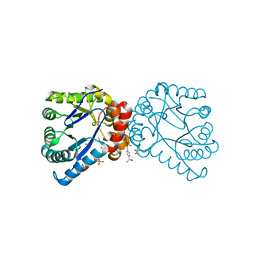 | |
4NIR
 
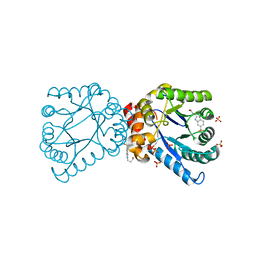 | |
3UYW
 
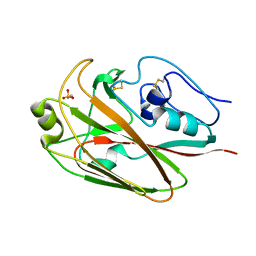 | | Crystal structures of globular head of 2009 pandemic H1N1 hemagglutinin | | Descriptor: | 2-AMINOETHANESULFONIC ACID, Hemagglutinin | | Authors: | Xuan, C.L, Shi, Y, Qi, J.X, Xiao, H.X, Gao, G.F. | | Deposit date: | 2011-12-06 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Structural vaccinology: structure-based design of influenza A virus hemagglutinin subtype-specific subunit vaccines
Protein Cell, 2, 2011
|
|
7XEE
 
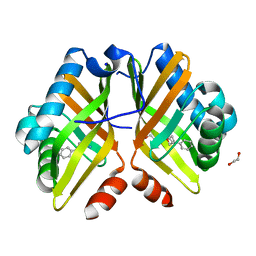 | | Crystal Structure of the Y53F/N55A mutant of LEH complexed with 2-(3-phenyloxetan-3-yl)ethanamine | | Descriptor: | 1,2-ETHANEDIOL, 2-(3-phenyloxetan-3-yl)ethanamine, Limonene-1,2-epoxide hydrolase, ... | | Authors: | Qu, G, Li, X, Sun, Z.T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.877 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
7XEF
 
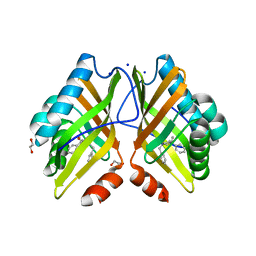 | | Crystal Structure of the Y53F/N55A mutant of LEH complexed with (R)-(1-benzyl-3-phenylpyrrolidin-3-yl)methanol | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Limonene-1,2-epoxide hydrolase, ... | | Authors: | Qu, G, Li, X, Sun, Z.T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
3UQR
 
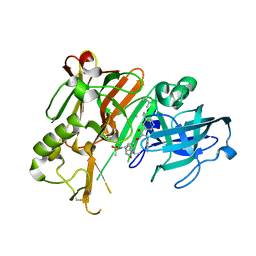 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, METHYL (2S)-1-[(2R,5S,8S,12S,13S)-2,13-DIBENZYL-12-HYDROXY-3,5-DIMETHYL-15-(3-[METHYL(METHYLSULFONYL)AMINO]-5-{[(1R)-1-PHENYLETHYL]CARBAMOYL}PHENYL)-8-(2-METHYLPROPYL)-4,7,10,15-TETRAOXO-3,6,9,14-TETRAAZAPENTADECAN-1-OYL]PYRROLIDINE-2-CARBOXYLATE | | Authors: | Chen, T.T, Chen, W.Y, Li, L, Xu, Y.C. | | Deposit date: | 2011-11-21 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.056 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
7Y57
 
 | |
4PBZ
 
 | | Structure of the human RbAp48-MTA1(670-695) complex | | Descriptor: | Histone-binding protein RBBP4, Metastasis-associated protein MTA1 | | Authors: | Murthy, A, Pei, X.Y, Watson, A.A, Silva, A.P.G, Mackay, J.P, Laue, E.D. | | Deposit date: | 2014-04-14 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Insight into the architecture of the NuRD complex: Structure of the RbAp48-MTA1 sub-complex.
J.Biol.Chem., 289, 2014
|
|
4JLW
 
 | | Crystal structure of formaldehyde dehydrogenase from Pseudomonas aeruginosa | | Descriptor: | Glutathione-independent formaldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Chen, S, Liao, Y.P, Wang, D.L, Wang, S, Ding, J.F, Wang, Y.M, Cai, L.J, Ran, X.Y, Zhu, H.X. | | Deposit date: | 2013-03-13 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of formaldehyde dehydrogenase from Pseudomonas aeruginosa: the binary complex with the cofactor NAD+.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4PBY
 
 | | Structure of the human RbAp48-MTA1(656-686) complex | | Descriptor: | Histone-binding protein RBBP4, ISOPROPYL ALCOHOL, Metastasis-associated protein MTA1 | | Authors: | Murthy, A, Lejon, S, Alqarni, S.S.M, Silva, A.P.G, Watson, A.A, Mackay, J.P, Laue, E.D. | | Deposit date: | 2014-04-14 | | Release date: | 2014-06-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into the architecture of the NuRD complex: Structure of the RbAp48-MTA1 sub-complex.
J.Biol.Chem., 289, 2014
|
|
3UYX
 
 | | Crystal structures of globular head of 2009 pandemic H1N1 hemagglutinin | | Descriptor: | Hemagglutinin, NITRATE ION | | Authors: | Xuan, C.L, Shi, Y, Qi, J.X, Xiao, H.X, Gao, G.F. | | Deposit date: | 2011-12-06 | | Release date: | 2012-10-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural vaccinology: structure-based design of influenza A virus hemagglutinin subtype-specific subunit vaccines
Protein Cell, 2, 2011
|
|
7YGI
 
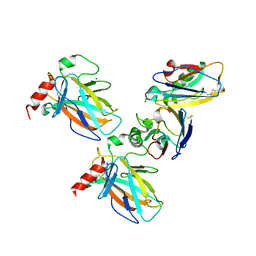 | | Crystal structure of p53 DBD domain in complex with azurin | | Descriptor: | Azurin, Cellular tumor antigen p53, PHOSPHATE ION, ... | | Authors: | Jiang, W.X, Zuo, J.Q, Hu, J.J, Chen, X.Q, Ma, L.X, Liu, Z, Xing, Q. | | Deposit date: | 2022-07-11 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of bacterial effector protein azurin targeting tumor suppressor p53 and inhibiting its ubiquitination.
Commun Biol, 6, 2023
|
|
3TCM
 
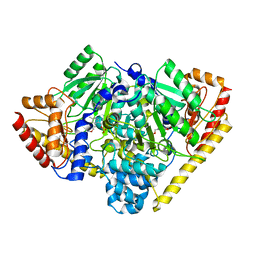 | | Crystal Structure of Alanine Aminotransferase from Hordeum vulgare | | Descriptor: | Alanine aminotransferase 2, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE | | Authors: | Rydel, T.J, Sturman, E.J, Halls, C, Chen, S, Zeng, J, Evdokimov, A, Duff, S.M.G. | | Deposit date: | 2011-08-09 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The Enzymology of alanine aminotransferase (AlaAT) isoforms from Hordeum vulgare and other organisms, and the HvAlaAT crystal structure.
Arch.Biochem.Biophys., 528, 2012
|
|
4PC0
 
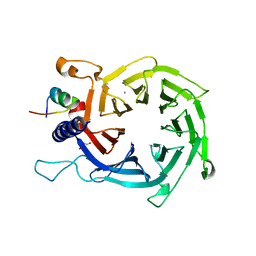 | | Structure of the human RbAp48-MTA1(670-711) complex | | Descriptor: | CALCIUM ION, GLYCEROL, Histone-binding protein RBBP4, ... | | Authors: | Alqarni, S.S.M, Silva, A.P.G, Mackay, J.P, Laue, E.D. | | Deposit date: | 2014-04-14 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into the architecture of the NuRD complex: Structure of the RbAp48-MTA1 sub-complex.
J.Biol.Chem., 289, 2014
|
|
7JGT
 
 | | Crystal Structure of FN3tt | | Descriptor: | Fibronectin type-III domain-containing protein | | Authors: | Luo, J, Malia, T.J. | | Deposit date: | 2020-07-19 | | Release date: | 2021-07-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Surface salt bridges contribute to the extreme thermal stability of an FN3-like domain from a thermophilic bacterium.
Proteins, 90, 2022
|
|
