1XCB
 
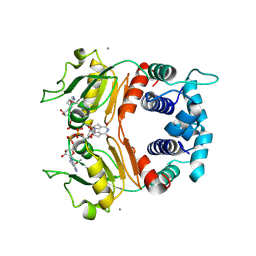 | | X-ray Structure of a Rex-Family Repressor/NADH Complex from Thermus Aquaticus | | Descriptor: | CALCIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Redox-sensing transcriptional repressor rex | | Authors: | Sickmier, E.A, Brekasis, D, Paranawithana, S, Bonanno, J.B, Burley, S.K, Paget, M.S, Kielkopf, C.L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-09-01 | | Release date: | 2004-09-28 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-Ray Structure of a Rex-Family Repressor/NADH Complex: Insights into the Mechanism of Redox Sensing
Structure, 13, 2005
|
|
2GUW
 
 | |
1QAJ
 
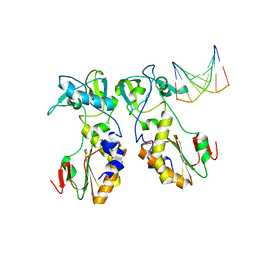 | | CRYSTAL STRUCTURES OF THE N-TERMINAL FRAGMENT FROM MOLONEY MURINE LEUKEMIA VIRUS REVERSE TRANSCRIPTASE COMPLEXED WITH NUCLEIC ACID: FUNCTIONAL IMPLICATIONS FOR TEMPLATE-PRIMER BINDING TO THE FINGERS DOMAIN | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*CP*AP*TP*G)-3'), REVERSE TRANSCRIPTASE | | Authors: | Najmudin, S, Cote, M, Sun, D, Yohannan, S, Montano, S.P, Gu, J, Georgiadis, M.M. | | Deposit date: | 1999-03-18 | | Release date: | 2000-04-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of an N-terminal fragment from Moloney murine leukemia virus reverse transcriptase complexed with nucleic acid: functional implications for template-primer binding to the fingers domain.
J.Mol.Biol., 296, 2000
|
|
1YBF
 
 | |
1Y9Q
 
 | |
1YDF
 
 | |
1YAV
 
 | |
1YBE
 
 | |
2I6E
 
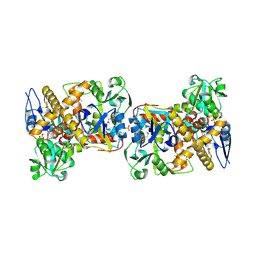 | | Crystal structure of protein DR0370 from Deinococcus radiodurans, Pfam DUF178 | | Descriptor: | Hypothetical protein, SULFATE ION | | Authors: | Tyagi, R, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-28 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structures of two proteins belonging to Pfam DUF178 revealed unexpected structural similarity to the DUF191 Pfam family.
Bmc Struct.Biol., 7, 2007
|
|
2I9U
 
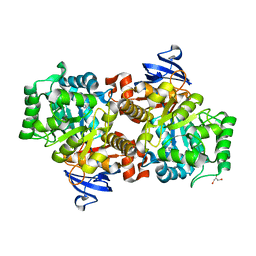 | |
1PXS
 
 | | Structure of Met56Ala mutant of Bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Faham, S, Yang, D, Bare, E, Yohannan, S, Whitelegge, J.P, Bowie, J.U. | | Deposit date: | 2003-07-06 | | Release date: | 2003-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Side-chain Contributions to Membrane Protein Structure and Stability.
J.Mol.Biol., 335, 2004
|
|
1YBD
 
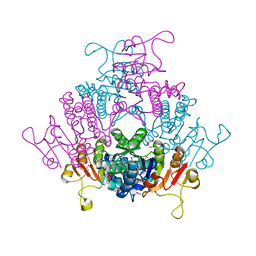 | |
1Y9I
 
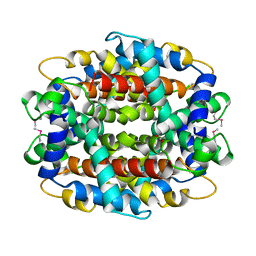 | | Crystal structure of low temperature requirement C protein from Listeria monocytogenes | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-15 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of phosphatidylglycerophosphatase (PGPase), a putative membrane-bound lipid phosphatase, reveals a novel binuclear metal binding site and two "proton wires".
Proteins, 64, 2006
|
|
1RKL
 
 | |
2NXO
 
 | |
2KN8
 
 | | NMR structure of the C-terminal domain of pUL89 | | Descriptor: | DNA cleavage and packaging protein large subunit, UL89 | | Authors: | Couvreux, A, Hantz, S, Marquant, R, Champier, G, Alain, S, Morellet, N, Bouaziz, S. | | Deposit date: | 2009-08-18 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insight into the structure of the pUL89 C-terminal domain of the human cytomegalovirus terminase complex.
Proteins, 78, 2010
|
|
2IMO
 
 | |
1YIR
 
 | |
1T61
 
 | | crystal structure of collagen IV NC1 domain from placenta basement membrane | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Vanacore, R.M, Shanmugasundararaj, S, Friedman, D.B, Bondar, O, Hudson, B.G, Sundaramoorthy, M. | | Deposit date: | 2004-05-05 | | Release date: | 2004-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The alpha1.alpha2 network of collagen IV. Reinforced stabilization of the noncollagenous domain-1 by noncovalent forces and the absence of Met-Lys cross-links
J.Biol.Chem., 279, 2004
|
|
1T3C
 
 | | Clostridium botulinum type E catalytic domain E212Q mutant | | Descriptor: | CHLORIDE ION, ZINC ION, neurotoxin type E | | Authors: | Agarwal, R, Eswaramoorthy, S, Kumaran, D, Binz, T, Swaminathan, S. | | Deposit date: | 2004-04-26 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of botulinum neurotoxin type E catalytic domain and its mutant Glu212-->Gln reveals the pivotal role of the Glu212 carboxylate in the catalytic pathway
Biochemistry, 43, 2004
|
|
2NYG
 
 | |
2IMG
 
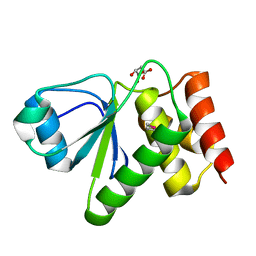 | | Crystal structure of dual specificity protein phosphatase 23 from Homo sapiens in complex with ligand malate ion | | Descriptor: | D-MALATE, Dual specificity protein phosphatase 23 | | Authors: | Agarwal, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-04 | | Release date: | 2006-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of human dual specificity protein phosphatase 23, VHZ, enzyme-substrate/product complex.
J.Biol.Chem., 283, 2008
|
|
1PXR
 
 | | Structure of Pro50Ala mutant of Bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Faham, S, Yang, D, Bare, E, Yohannan, S, Whitelegge, J.P, Bowie, J.U. | | Deposit date: | 2003-07-06 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Side-chain Contributions to Membrane Protein Structure and Stability.
J.Mol.Biol., 335, 2004
|
|
1PY6
 
 | | Bacteriorhodopsin crystallized from bicells | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Faham, S, Yang, D, Bare, E, Yohannan, S, Whitelegge, J.P, Bowie, J.U. | | Deposit date: | 2003-07-08 | | Release date: | 2003-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Side-chain Contributions to Membrane Protein Structure and Stability.
J.Mol.Biol., 335, 2004
|
|
2M54
 
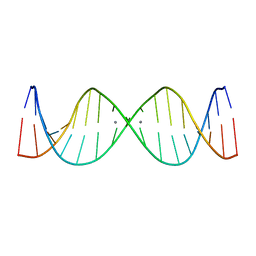 | | Refined NMR solution structure of metal-modified DNA | | Descriptor: | DNA (5'-D(*TP*TP*AP*AP*TP*TP*TP*(D33)P*(D33)P*(D33)P*AP*AP*AP*TP*TP*AP*A)-3'), SILVER ION | | Authors: | Kumbhar, S, Johannsen, S, Sigel, R.K, Waller, M.P, Mueller, J. | | Deposit date: | 2013-02-13 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A QM/MM refinement of an experimental DNA structure with metal-mediated base pairs.
J.Inorg.Biochem., 127, 2013
|
|
