4IV3
 
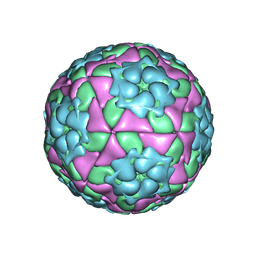 | | Crystal structure of recombinant foot-and-mouth-disease virus A22-H2093C empty capsid | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Porta, C, Kotecha, A, Burman, A, Jackson, T, Ren, J, Loureiro, S, Jones, I.M, Fry, E.E, Stuart, D.I, Charleston, B. | | Deposit date: | 2013-01-22 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Rational engineering of recombinant picornavirus capsids to produce safe, protective vaccine antigen.
Plos Pathog., 9, 2013
|
|
4J0S
 
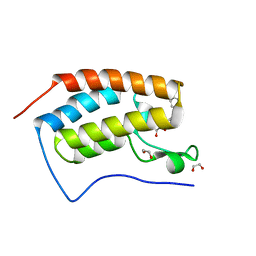 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(S)-hydroxy(phenyl)methyl]phenol, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, Hewings, D.S, von Delft, F, Conway, S.J, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-01-31 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Optimization of 3,5-dimethylisoxazole derivatives as potent bromodomain ligands.
J.Med.Chem., 56, 2013
|
|
6MH8
 
 | | High-viscosity injector-based Pink Beam Serial Crystallography of Micro-crystals at a Synchrotron Radiation Source | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a, Soluble cytochrome b562 chimeric construct | | Authors: | Martin-Garcia, J.M, Zhu, L, Mendez, D, Lee, M, Chun, E, Li, C, Hu, H, Subramanian, G, Kissick, D, Ogata, C, Henning, R, Ishchenko, A, Dobson, Z, Zhan, S, Weierstall, U, Spence, J.C.H, Fromme, P, Zatsepin, N.A, Fischetti, R.F, Cherezov, V, Liu, W. | | Deposit date: | 2018-09-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | High-viscosity injector-based pink-beam serial crystallography of microcrystals at a synchrotron radiation source.
Iucrj, 6, 2019
|
|
4NR9
 
 | | Crystal Structure of the bromodomain of human BAZ2B in complex with acetylated lysine | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, N(6)-ACETYLLYSINE | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Targeting low-druggability bromodomains: fragment based screening and inhibitor design against the BAZ2B bromodomain.
J.Med.Chem., 56, 2013
|
|
6MKJ
 
 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the closed conformation | | Descriptor: | penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Soares, A, D'Andrea, E.D, Jaconcic, J, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.864 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
4IW0
 
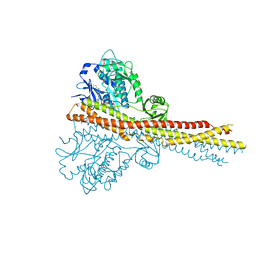 | | Crystal structure and mechanism of activation of TBK1 | | Descriptor: | N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Larabi, A, Devos, J.M, Ng, S.-L, Nanao, M.H, Round, A, Maniatis, T, Panne, D. | | Deposit date: | 2013-01-23 | | Release date: | 2013-03-13 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structure and mechanism of activation of TANK-binding kinase 1.
Cell Rep, 3, 2013
|
|
2YEY
 
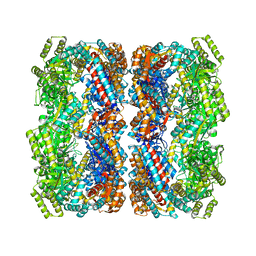 | | Crystal structure of the allosteric-defective chaperonin GroEL E434K mutant | | Descriptor: | 60 KDA CHAPERONIN | | Authors: | Cabo-Bilbao, A, Mechaly, A.E, Agirre, J, Spinelli, S, Sot, B, Muga, A, Guerin, D.M.A. | | Deposit date: | 2011-03-31 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Crystal Structure of the Temperature-Sensitive and Allosteric-Defective Chaperonin Groele461K.
J.Struct.Biol., 155, 2006
|
|
6MKV
 
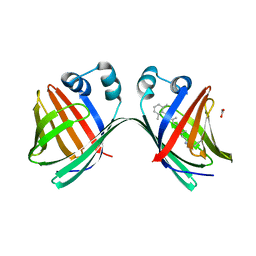 | |
2YJ7
 
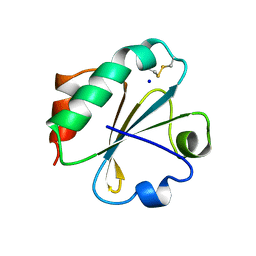 | | Crystal structure of a hyperstable protein from the Precambrian period | | Descriptor: | LPBCA THIOREDOXIN, SODIUM ION | | Authors: | Gavira, J.A, Ingles, A, Ibarra, B, Garcia-Ruiz, J.M. | | Deposit date: | 2011-05-19 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Conservation of Protein Structure Over Four Billion Years
Structure, 21, 2013
|
|
3FEO
 
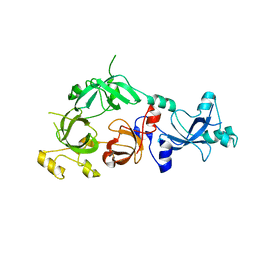 | | The crystal structure of MBTD1 | | Descriptor: | MBT domain-containing protein 1 | | Authors: | Amaya, M.F, Eryilmaz, J, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-30 | | Release date: | 2009-01-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies of a four-MBT repeat protein MBTD1.
Plos One, 4, 2009
|
|
4NV8
 
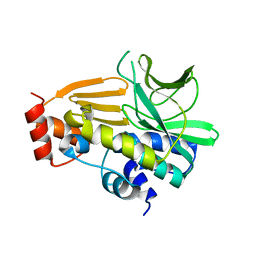 | | Crystal Structure of Mesorhizobium Loti Arylamine N-acetyltransferase F42W Mutant | | Descriptor: | Arylamine N-acetyltransferase | | Authors: | Xu, X.M, Haouz, A, Weber, P, Li de la sierra-gallay, I, Kubiak, X, Dupret, J.-M, Rodrigues-lima, F. | | Deposit date: | 2013-12-05 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Insight into cofactor recognition in arylamine N-acetyltransferase enzymes: structure of Mesorhizobium loti arylamine N-acetyltransferase in complex with coenzyme A.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7WUX
 
 | | Crystal structure of AziU3/U2 complexed with (5S,6S)-O7-sulfo DADH from Streptomyces sahachiroi | | Descriptor: | (2S,5S,6S)-2,6-bis(azanyl)-5-oxidanyl-7-sulfooxy-heptanoic acid, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, AziU2, ... | | Authors: | Kurosawa, S, Yoshida, A, Tomita, T, Nishiyama, M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Enzymatic Aziridine Formation via Sulfate Elimination.
J.Am.Chem.Soc., 144, 2022
|
|
1XDU
 
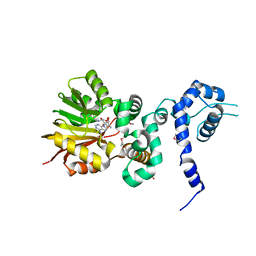 | | Crystal structure of Aclacinomycin-10-hydroxylase (RdmB) in complex with Sinefungin (SFG) | | Descriptor: | ACETATE ION, Protein RdmB, SINEFUNGIN | | Authors: | Jansson, A, Koskiniemi, H, Erola, A, Wang, J, Mantsala, P, Schneider, G, Niemi, J. | | Deposit date: | 2004-09-08 | | Release date: | 2004-11-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aclacinomycin 10-Hydroxylase Is a Novel Substrate-assisted Hydroxylase Requiring S-Adenosyl-L-methionine as Cofactor
J.Biol.Chem., 280, 2005
|
|
6ETN
 
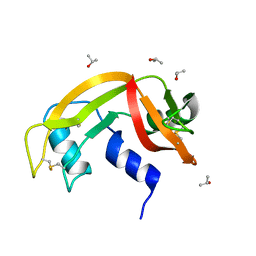 | |
1FH5
 
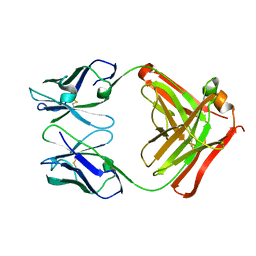 | | CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF THE MONOCLONAL ANTIBODY MAK33 | | Descriptor: | MONOCLONAL ANTIBODY MAK33 | | Authors: | Augustine, J.G, de la Calle, A, Knarr, G, Buchner, J, Frederick, C.A. | | Deposit date: | 2000-07-31 | | Release date: | 2000-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of the fab fragment of the monoclonal antibody MAK33. Implications for folding and interaction with the chaperone bip.
J.Biol.Chem., 276, 2001
|
|
4IWS
 
 | | Putative Aromatic Acid Decarboxylase | | Descriptor: | PA0254, SULFATE ION | | Authors: | Jacewicz, A, Izumi, A, Brunner, K, Schneider, G. | | Deposit date: | 2013-01-24 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into the UbiD Protein Family from the Crystal Structure of PA0254 from Pseudomonas aeruginosa.
Plos One, 8, 2013
|
|
1XEU
 
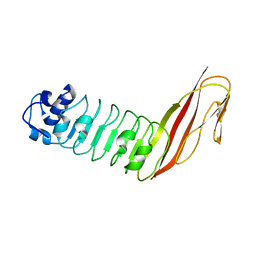 | | Crystal Structure of Internalin C from Listeria monocytogenes | | Descriptor: | internalin C | | Authors: | Ooi, A, Hussain, S, Seyedarabi, A, Pickersgill, R.W. | | Deposit date: | 2004-09-13 | | Release date: | 2005-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of internalin C from Listeria monocytogenes.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2YNH
 
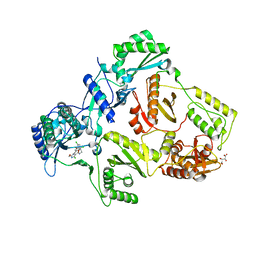 | | HIV-1 Reverse Transcriptase in complex with inhibitor GSK500 | | Descriptor: | 4-chloranyl-N-[[4-chloranyl-3-(3-chloranyl-5-cyano-phenoxy)-2-fluoranyl-phenyl]methyl]-2-(hydroxymethyl)-1H-imidazole-5-carboxamide, D(-)-TARTARIC ACID, P51 RT, ... | | Authors: | Chong, P, Sebahar, P, Youngman, M, Garrido, D, Zhang, H, Stewart, E.L, Nolte, R.T, Wang, L, Ferris, R.G, Edelstein, M, Weaver, K, Mathis, A, Peat, A. | | Deposit date: | 2012-10-14 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Rational Design of Potent Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase.
J.Med.Chem., 55, 2012
|
|
5MWA
 
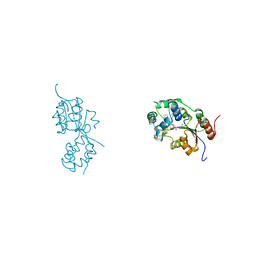 | | human sEH Phosphatase in complex with 3-4-3,4-dichlorophenyl-5-phenyl-1,3-oxazol-2-yl-benzoic-acid | | Descriptor: | 3-[4-(3,4-dichlorophenyl)-5-phenyl-1,3-oxazol-2-yl]benzoic acid, Bifunctional epoxide hydrolase 2, MAGNESIUM ION | | Authors: | Kramer, J.S, Pogoryelov, D, Sorrell, F.J, Fox, N, Chaikuad, A, Knapp, S, Proschak, E. | | Deposit date: | 2017-01-18 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of first in vivo active inhibitors of soluble epoxide hydrolase (sEH) phosphatase domain.
J.Med.Chem., 2019
|
|
7WUW
 
 | | Crystal structure of AziU3/U2 from Streptomyces sahachiroi | | Descriptor: | AziU2, AziU3, MAGNESIUM ION, ... | | Authors: | Kurosawa, S, Yoshida, A, Tomita, T, Nishiyama, M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular Basis for Enzymatic Aziridine Formation via Sulfate Elimination.
J.Am.Chem.Soc., 144, 2022
|
|
7XGO
 
 | | Human renin in complex with compound2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin, UNKNOWN LIGAND | | Authors: | Kashima, A. | | Deposit date: | 2022-04-05 | | Release date: | 2022-08-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Novel 2-Carbamoyl Morpholine Derivatives as Highly Potent and Orally Active Direct Renin Inhibitors.
Acs Med.Chem.Lett., 13, 2022
|
|
5MT4
 
 | |
3EYA
 
 | | Structural basis for membrane binding and catalytic activation of the peripheral membrane enzyme pyruvate oxidase from Escherichia coli | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Neumann, P, Weidner, A, Pech, A, Stubbs, M.T, Tittmann, K. | | Deposit date: | 2008-10-20 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for membrane binding and catalytic activation of the peripheral membrane enzyme pyruvate oxidase from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2VWA
 
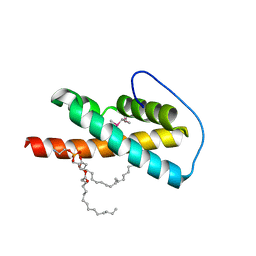 | | Crystal structure of a sporozoite protein essential for liver stage development of malaria parasite | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, PUTATIVE UNCHARACTERIZED PROTEIN PF13_0012 | | Authors: | Sharma, A, Sharma, A, Yogavel, M, Gill, J, Akhouri, R.R. | | Deposit date: | 2008-06-20 | | Release date: | 2008-07-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Soluble Domain of Malaria Sporozoite Protein Uis3 in Complex with Lipid.
J.Biol.Chem., 283, 2008
|
|
2YNG
 
 | | HIV-1 Reverse Transcriptase in complex with inhibitor GSK560 | | Descriptor: | 2-azanyl-N-[[4-bromanyl-3-(3-chloranyl-5-cyano-phenoxy)-2-fluoranyl-phenyl]methyl]-4-chloranyl-1H-imidazole-5-carboxamide, MAGNESIUM ION, P51 RT, ... | | Authors: | Chong, P, Sebahar, P, Youngman, M, Garrido, D, Zhang, H, Stewart, E.L, Nolte, R.T, Wang, L, Ferris, R.G, Edelstein, M, Weaver, K, Mathis, A, Peat, A. | | Deposit date: | 2012-10-14 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Rational Design of Potent Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase.
J.Med.Chem., 55, 2012
|
|
