1VCH
 
 | |
1VE4
 
 | |
1VDK
 
 | |
1UJV
 
 | |
1UJY
 
 | | Solution structure of SH3 domain in Rac/Cdc42 guanine nucleotide exchange factor(GEF) 6 | | Descriptor: | Rho guanine nucleotide exchange factor 6 | | Authors: | He, F, Muto, Y, Uda, H, Koshiba, S, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nagayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-12 | | Release date: | 2004-02-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SH3 domain in Rac/Cdc42 guanine nucleotide exchange factor(GEF) 6
To be Published
|
|
1UJU
 
 | |
1VC8
 
 | |
1UKK
 
 | | Structure of Osmotically Inducible Protein C from Thermus thermophilus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Osmotically Inducible Protein C | | Authors: | Rehse, P.H, Kuramitsu, S, Yokoyama, S, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-24 | | Release date: | 2004-05-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Structure and Biochemical Analysis of the Thermus thermophilus Osmotically Inducible Protein C
J.MOL.BIOL., 338, 2004
|
|
1UKW
 
 | | Crystal structure of medium-chain acyl-CoA dehydrogenase from Thermus thermophilus HB8 | | Descriptor: | COBALT (II) ION, FLAVIN-ADENINE DINUCLEOTIDE, acyl-CoA dehydrogenase | | Authors: | Hamada, K, Ago, H, Kuramitsu, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-02 | | Release date: | 2004-11-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Thermus thermophilus medium-chain acyl-CoA dehydrogenase
To be published
|
|
1UK5
 
 | | Solution structure of the Murine BAG domain of Bcl2-associated athanogene 3 | | Descriptor: | BAG-family molecular chaperone regulator-3 | | Authors: | Hatta, R, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-19 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The C-terminal BAG domain of BAG5 induces conformational changes of the Hsp70 nucleotide-binding domain for ADP-ATP exchange
Structure, 18, 2010
|
|
2RQ4
 
 | | Refinement of RNA binding domain 3 in CUG triplet repeat RNA-binding protein 1 | | Descriptor: | CUG-BP- and ETR-3-like factor 1 | | Authors: | Tsuda, K, Kuwasako, K, Takahashi, M, Someya, T, Inoue, M, Terada, T, Kobayashi, N, Shirouzu, M, Kigawa, T, Guntert, P, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-01-19 | | Release date: | 2009-08-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3
Nucleic Acids Res., 2009
|
|
1VBI
 
 | |
1UM7
 
 | |
1V6G
 
 | | Solution Structure of the LIM Domain of the Human Actin Binding LIM Protein 2 | | Descriptor: | Actin Binding LIM Protein 2, ZINC ION | | Authors: | Miyamoto, K, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-29 | | Release date: | 2004-06-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the LIM Domain of the Human Actin Binding LIM Protein 2
To be Published
|
|
1V95
 
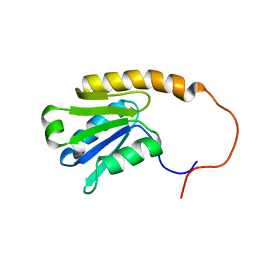 | | Solution Structure of Anticodon Binding Domain from Nuclear Receptor Coactivator 5 (Human KIAA1637 Protein) | | Descriptor: | Nuclear receptor coactivator 5 | | Authors: | Zhao, C, Kigawa, T, Tochio, N, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-20 | | Release date: | 2004-07-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Anticodon Binding Domain from Nuclear Receptor Coactivator 5 (Human KIAA1637 Protein)
To be Published
|
|
1VCD
 
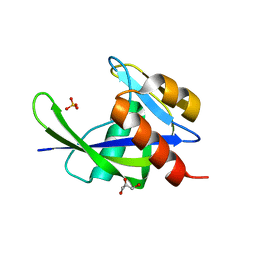 | | Crystal Structure of a T.thermophilus HB8 Ap6A hydrolase Ndx1 | | Descriptor: | GLYCEROL, Ndx1, SULFATE ION | | Authors: | Iwai, T, Nakagawa, N, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-05 | | Release date: | 2005-04-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Nudix Protein Ndx1 from Thermus thermophilus HB8 in binary complex with diadenosine hexaphosphate
To be Published
|
|
1V2D
 
 | |
1V67
 
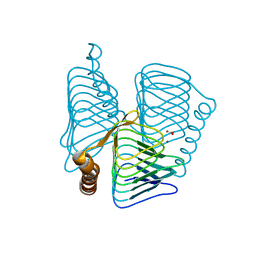 | | Structure of ferripyochelin binding protein from pyrococcus horikoshii OT3 | | Descriptor: | BICARBONATE ION, CALCIUM ION, ZINC ION, ... | | Authors: | Jeyakanthan, J, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-27 | | Release date: | 2003-12-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Observation of a calcium-binding site in the gamma-class carbonic anhydrase from Pyrococcus horikoshii.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1VC9
 
 | | Crystal Structure of a T.thermophilus HB8 Ap6A hydrolase E50Q mutant-Mg2+-ATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ndx1 | | Authors: | Iwai, T, Nakagawa, N, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-05 | | Release date: | 2005-04-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Nudix Protein Ndx1 from Thermus thermophilus HB8 in binary complex with diadenosine hexaphosphate
To be Published
|
|
1V6S
 
 | |
1V8F
 
 | |
1V38
 
 | | Solution structure of the Sterile Alpha Motif (SAM) domain of mouse SAMSN1 | | Descriptor: | SAM-domain protein SAMSN-1 | | Authors: | Goroncy, A, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-29 | | Release date: | 2004-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Sterile Alpha Motif (SAM) domain of mouse SAMSN1
To be Published
|
|
1V4P
 
 | |
1V7Q
 
 | |
1VCF
 
 | | Crystal Structure of IPP isomerase at I422 | | Descriptor: | CADMIUM ION, FLAVIN MONONUCLEOTIDE, isopentenyl-diphosphate delta-isomerase | | Authors: | Wada, T, Park, S.-Y, Tame, R.H, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-08 | | Release date: | 2005-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of IPP isomerase at I422
To be Published
|
|
