8UYE
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 1 | | Descriptor: | BtCoV-HKU5 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYP
 
 | | SARS-CoV-1 5' proximal stem-loop 5 | | Descriptor: | SARS-CoV-1 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7Y5G
 
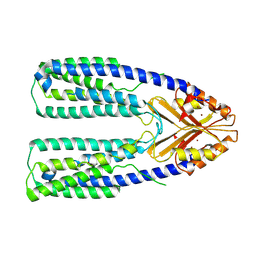 | | Cryo-EM structure of a eukaryotic ZnT8 in the presence of zinc | | Descriptor: | ZINC ION, Zinc transporter 8 | | Authors: | Zhang, S, Fu, C, Luo, Y, Sun, Z, Su, Z, Zhou, X. | | Deposit date: | 2022-06-17 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Cryo-EM structure of a eukaryotic zinc transporter at a low pH suggests its Zn 2+ -releasing mechanism.
J.Struct.Biol., 215, 2022
|
|
7Y5H
 
 | | Cryo-EM structure of a eukaryotic ZnT8 at a low pH | | Descriptor: | ZINC ION, Zinc transporter 8 | | Authors: | Zhang, S, Fu, C, Luo, Y, Sun, Z, Su, Z, Zhou, X. | | Deposit date: | 2022-06-17 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Cryo-EM structure of a eukaryotic zinc transporter at a low pH suggests its Zn 2+ -releasing mechanism.
J.Struct.Biol., 215, 2022
|
|
8W4C
 
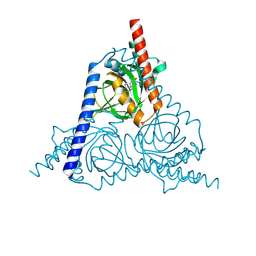 | | The sigma-1 receptor from Xenopus laevis in complex with progesterone by soaking | | Descriptor: | PROGESTERONE, Sigma non-opioid intracellular receptor 1 | | Authors: | Xiao, Y, Fu, C, Sun, Z, Zhou, X. | | Deposit date: | 2023-08-23 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.676 Å) | | Cite: | Insight into binding of endogenous neurosteroid ligands to the sigma-1 receptor.
Nat Commun, 15, 2024
|
|
8W4B
 
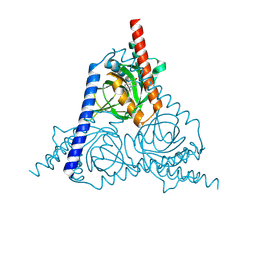 | | The sigma-1 receptor from Xenopus laevis in complex with progesterone by co-crystallization | | Descriptor: | PROGESTERONE, Sigma non-opioid intracellular receptor 1 | | Authors: | Xiao, Y, Fu, C, Sun, Z, Zhou, X. | | Deposit date: | 2023-08-23 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Insight into binding of endogenous neurosteroid ligands to the sigma-1 receptor.
Nat Commun, 15, 2024
|
|
8W4D
 
 | |
8W4E
 
 | |
7D5I
 
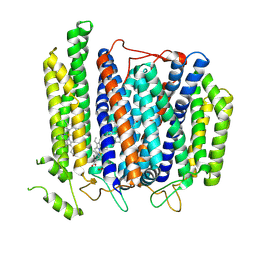 | | Structure of Mycobacterium smegmatis bd complex in the apo-form. | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome D ubiquinol oxidase subunit 1, HEME B/C, ... | | Authors: | Wang, W, Gong, H, Gao, Y, Zhou, X, Rao, Z. | | Deposit date: | 2020-09-26 | | Release date: | 2021-06-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structure of mycobacterial cytochrome bd reveals two oxygen access channels.
Nat Commun, 12, 2021
|
|
8VBI
 
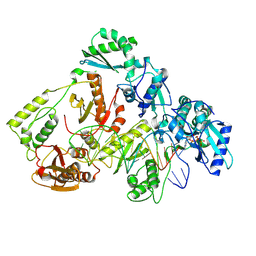 | | Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (38-MER), HIV-1 reverse transcriptase P51 subunit, ... | | Authors: | Vergara, S, Zhou, X, Santiago, U, Conway, J.F, Sluis-Cremer, N, Calero, G. | | Deposit date: | 2023-12-12 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis of deoxynucleotide addition by HIV-1 RT during reverse transcription.
Nat Commun, 15, 2024
|
|
8VBE
 
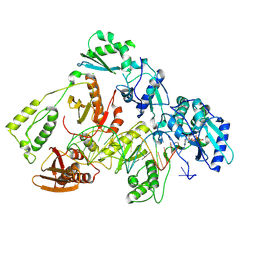 | | Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (39-MER), HIV-1 reverse transcriptase P51 subunit, ... | | Authors: | Vergara, S, Zhou, X, Santiago, U, Conway, J.F, Sluis-Cremer, N, Calero, G. | | Deposit date: | 2023-12-12 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of deoxynucleotide addition by HIV-1 RT during reverse transcription.
Nat Commun, 15, 2024
|
|
8VBF
 
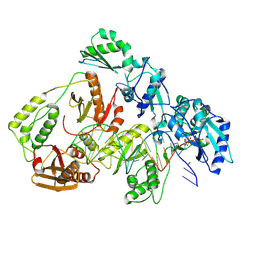 | | Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (40-MER), HIV-1 reverse transcriptase P51 subunit, ... | | Authors: | Vergara, S, Zhou, X, Santiago, U, Conway, J.F, Sluis-Cremer, N, Calero, G. | | Deposit date: | 2023-12-12 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of deoxynucleotide addition by HIV-1 RT during reverse transcription.
Nat Commun, 15, 2024
|
|
8VBH
 
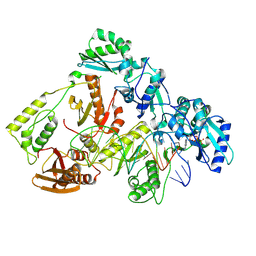 | | Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (38-MER), HIV-1 reverse transcriptase P51 subunit, ... | | Authors: | Vergara, S, Zhou, X, Santiago, U, Conway, J.F, Sluis-Cremer, N, Calero, G. | | Deposit date: | 2023-12-12 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural basis of deoxynucleotide addition by HIV-1 RT during reverse transcription.
Nat Commun, 15, 2024
|
|
8VBG
 
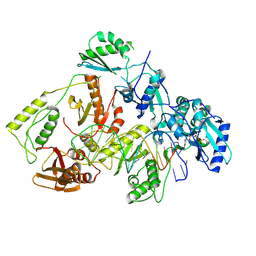 | | Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM | | Descriptor: | 2'-deoxy-5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]adenosine, DNA (38-MER), HIV-1 reverse transcriptase P51 subunit, ... | | Authors: | Vergara, S, Zhou, X, Santiago, U, Conway, J.F, Sluis-Cremer, N, Calero, G. | | Deposit date: | 2023-12-12 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of deoxynucleotide addition by HIV-1 RT during reverse transcription.
Nat Commun, 15, 2024
|
|
8VB9
 
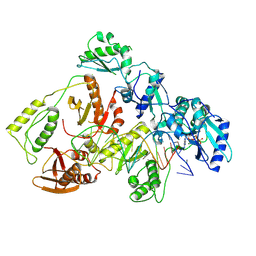 | | Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (38-MER), HIV-1 reverse transcriptase P51 subunit, ... | | Authors: | Vergara, S, Zhou, X, Santiago, U, Conway, J.F, Sluis-Cremer, N, Calero, G. | | Deposit date: | 2023-12-12 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of deoxynucleotide addition by HIV-1 RT during reverse transcription.
Nat Commun, 15, 2024
|
|
8VB8
 
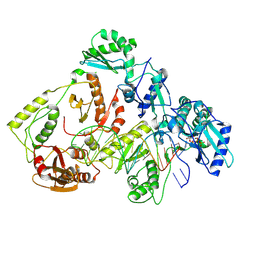 | | Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (38-MER), HIV-1 reverse transcriptase P51 subunit, ... | | Authors: | Vergara, S, Zhou, X, Santiago, U, Conway, J.F, Sluis-Cremer, N, Calero, G. | | Deposit date: | 2023-12-12 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of deoxynucleotide addition by HIV-1 RT during reverse transcription.
Nat Commun, 15, 2024
|
|
8VBC
 
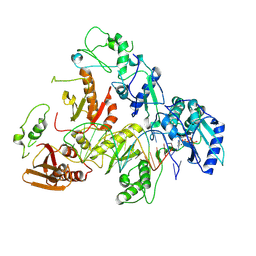 | | Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (38-MER), HIV-1 reverse transcriptase P51 subunit, ... | | Authors: | Vergara, S, Zhou, X, Santiago, U, Conway, J.F, Sluis-Cremer, N, Calero, G. | | Deposit date: | 2023-12-12 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of deoxynucleotide addition by HIV-1 RT during reverse transcription.
Nat Commun, 15, 2024
|
|
8VB6
 
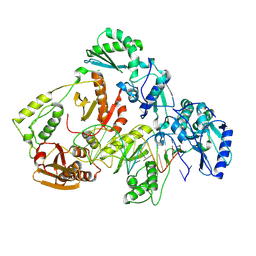 | | Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM | | Descriptor: | DNA (38-MER), HIV-1 reverse transcriptase P51 subunit, HIV-1 reverse transcriptase/ribonuclease H P66 subunit | | Authors: | Vergara, S, Zhou, X, Santiago, U, Conway, J.F, Sluis-Cremer, N, Calero, G. | | Deposit date: | 2023-12-12 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of deoxynucleotide addition by HIV-1 RT during reverse transcription.
Nat Commun, 15, 2024
|
|
8VB7
 
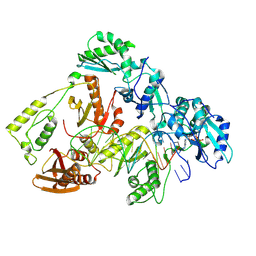 | | Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (38-MER), HIV-1 reverse transcriptase P51 subunit, ... | | Authors: | Vergara, S, Zhou, X, Santiago, U, Conway, J.F, Sluis-Cremer, N, Calero, G. | | Deposit date: | 2023-12-12 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis of deoxynucleotide addition by HIV-1 RT during reverse transcription.
Nat Commun, 15, 2024
|
|
8VBD
 
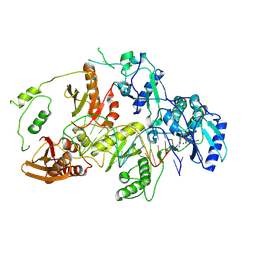 | | Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (38-MER), HIV-1 reverse transcriptase P51 subunit, ... | | Authors: | Vergara, S, Zhou, X, Santiago, U, Conway, J.F, Sluis-Cremer, N, Calero, G. | | Deposit date: | 2023-12-12 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of deoxynucleotide addition by HIV-1 RT during reverse transcription.
Nat Commun, 15, 2024
|
|
8YBB
 
 | |
8XIU
 
 | | Cryo-EM structure of a frog VMAT2 in an apo conformation | | Descriptor: | XlVMAT2 | | Authors: | Lyu, Y, Fu, C, Ma, H, Sun, Z, Su, Z, Zhou, X. | | Deposit date: | 2023-12-20 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Engineering of a mammalian VMAT2 for cryo-EM analysis results in non-canonical protein folding.
Nat Commun, 15, 2024
|
|
8XIT
 
 | | Cryo-EM structure of sheep VMAT2 dimer in an atypical fold | | Descriptor: | OaVMAT2-BRIL | | Authors: | Lyu, Y, Fu, C, Ma, H, Sun, Z, Su, Z, Zhou, X. | | Deposit date: | 2023-12-20 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Engineering of a mammalian VMAT2 for cryo-EM analysis results in non-canonical protein folding.
Nat Commun, 15, 2024
|
|
8WUE
 
 | |
8WWB
 
 | |
