8JNK
 
 | |
8JNR
 
 | |
8AHX
 
 | |
2P87
 
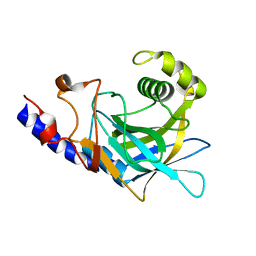 | | Crystal structure of the C-terminal domain of C. elegans pre-mRNA splicing factor Prp8 | | Descriptor: | Pre-mRNA-splicing factor Prp8 | | Authors: | Zhang, L, Shen, J, Guarnieri, M.T, Heroux, A, Yang, K, Zhao, R. | | Deposit date: | 2007-03-21 | | Release date: | 2007-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the C-terminal domain of splicing factor Prp8 carrying retinitis pigmentosa mutants
Protein Sci., 16, 2007
|
|
3QAU
 
 | | 3-Hydroxy-3-MethylGlutaryl-Coenzyme A Reductase from Streptococcus pneumoniae | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme a reductase, GLYCEROL, SODIUM ION, ... | | Authors: | Zhang, L, Feng, L, Zhou, L, Gui, J, Wan, J. | | Deposit date: | 2011-01-11 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-Hydroxy-3-MethylGlutaryl-Coenzyme A Reductase from Streptococcus pneumoniae
To be Published
|
|
8JKK
 
 | |
6IPB
 
 | |
7CLZ
 
 | |
7XMK
 
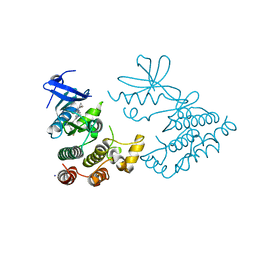 | | Crystal structure of human RIPK1 kinase domain in complex with compound SKLB923 | | Descriptor: | 5-[2-(cyclopropylcarbonylamino)-[1,2,4]triazolo[1,5-a]pyridin-7-yl]-N-[(1S)-1-(3-fluorophenyl)ethyl]-1-methyl-indole-3-carboxamide, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Zhang, L, Wang, Y, Li, Y, Yang, S. | | Deposit date: | 2022-04-26 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.376 Å) | | Cite: | From Hit to Lead: Structure-Based Optimization of Novel Selective Inhibitors of Receptor-Interacting Protein Kinase 1 (RIPK1) for the Treatment of Inflammatory Diseases.
J.Med.Chem., 67, 2024
|
|
7VTY
 
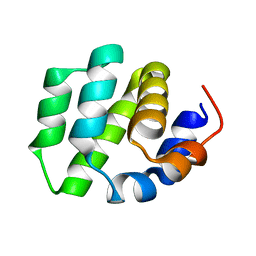 | | de novo designed protein | | Descriptor: | de novo designed protein | | Authors: | Zhang, L. | | Deposit date: | 2021-10-31 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rotamer-free protein sequence design based on deep learning and self-consistency.
Nat Comput Sci, 2023
|
|
7VQV
 
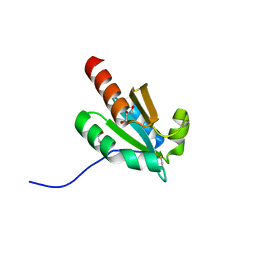 | | de novo design based on 1r26 | | Descriptor: | GLYCEROL, de novo designed protein | | Authors: | Zhang, L. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Rotamer-free protein sequence design based on deep learning and self-consistency.
Nat Comput Sci, 2023
|
|
7VU4
 
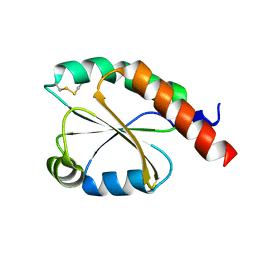 | | de novo design based on 1r26 | | Descriptor: | de novo design protein | | Authors: | Zhang, L. | | Deposit date: | 2021-11-01 | | Release date: | 2022-06-08 | | Last modified: | 2023-06-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rotamer-free protein sequence design based on deep learning and self-consistency.
Nat Comput Sci, 2023
|
|
7VQL
 
 | | de novo designed based on 1r26 | | Descriptor: | AMMONIUM ION, GLYCEROL, de novo designed protein | | Authors: | Zhang, L. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rotamer-free protein sequence design based on deep learning and self-consistency.
Nat Comput Sci, 2023
|
|
7VQW
 
 | | de novo designed protein based on 1r26 | | Descriptor: | de novo designed protein | | Authors: | Zhang, L. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Rotamer-free protein sequence design based on deep learning and self-consistency.
Nat Comput Sci, 2023
|
|
7C5G
 
 | | Crystal Structure of C150S mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with PO4 at 1.98 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5N
 
 | | Crystal Structure of C150A+H177A mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli at 2.0 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5H
 
 | | Crystal Structure of Glyceraldehyde-3-phosphate dehydrogenase1 from Escherichia coli at 2.09 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5R
 
 | | Crystal Structure of C150S mutant Glyceraldehyde-3-phosphate dehydrogenase1 from Escherichia coli complexed with BPG at 2.31 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCERALDEHYDE-3-PHOSPHATE, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5I
 
 | | Crystal Structure of C150A mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with PO4 at 2.49 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5Q
 
 | | Crystal Structure of H177A mutant Glyceraldehyde-3-phosphate dehydrogenase1 from Escherichia coli complexed with BPG at 2.13 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCERALDEHYDE-3-PHOSPHATE, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5K
 
 | | Crystal Structure of C150S mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with G3P at 2.69 Angstrom resolution | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, GLYCERALDEHYDE-3-PHOSPHATE, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5L
 
 | | Crystal Structure of C150S mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli at 2.1 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5P
 
 | | Crystal Structure Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with G3P at 2.35 Angstrom resolution. | | Descriptor: | CHLORIDE ION, GLYCERALDEHYDE-3-PHOSPHATE, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5J
 
 | | Crystal Structure of C150A mutant of Glyceraldehyde-3-phosphate dehydrogenase1 from Escherichia coli at 1.98 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C7K
 
 | | Crystal Structure of Thioacyl-Glyceraldehyde-3-phosphate dehydrogenase 1(GAPDH 1) from Escherichia coli at 1.77 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, ACETONE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-26 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
