9FX7
 
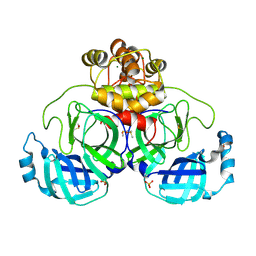 | | Crystal structure of Cryo2RT SARS-CoV-2 main protease at 294K | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Huang, C.Y, Aumonier, S, Mac Sweeney, A, Olieric, V, Wang, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
9FX4
 
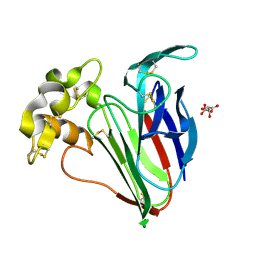 | | Crystal structure of Cryo2RT Thaumatin at 100K | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Huang, C.Y, Aumonier, S, Olieric, V, Wang, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
2P09
 
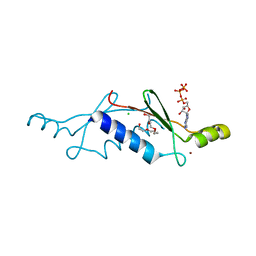 | | Structural Insights into the Evolution of a Non-Biological Protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Smith, M, Rosenow, M, Wang, M, Allen, J.P, Szostak, J.W, Chaput, J.C. | | Deposit date: | 2007-02-28 | | Release date: | 2007-06-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into the evolution of a non-biological protein: importance of surface residues in protein fold optimization.
PLoS ONE, 2, 2007
|
|
6ESJ
 
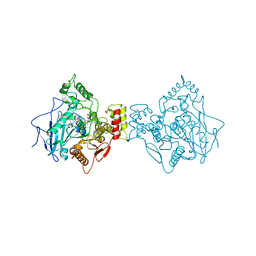 | | Human butyrylcholinesterase in complex with propidium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3,8-DIAMINO-5[3-(DIETHYLMETHYLAMMONIO)PROPYL]-6-PHENYLPHENANTHRIDINIUM, ... | | Authors: | Nachon, F, Brazzolotto, X, Wandhammer, M, Trovaslet-Leroy, M, Macdonald, I.R, Darvesh, S, Rosenberry, T.L. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.979845 Å) | | Cite: | Comparison of the Binding of Reversible Inhibitors to Human Butyrylcholinesterase and Acetylcholinesterase: A Crystallographic, Kinetic and Calorimetric Study.
Molecules, 22, 2017
|
|
8XT3
 
 | | Cryo-EM structure of the human 39S mitoribosome with 10uM Tigecycline | | Descriptor: | 16s rRNA, 39S ribosomal protein L22, mitochondrial, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8XSX
 
 | | Cryo-EM structure of the human 80S ribosome with Tigecycline, E-tRNA, SERBP1 and eEF2 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8XT0
 
 | | Cryo-EM structure of the human 55S mitoribosome with 5um Tigecycline | | Descriptor: | 12s rRNA, 16s rRNA, 39S ribosomal protein L22, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8XSZ
 
 | | Cryo-EM structure of the human 80S ribosome with Tigecycline, E-tRNA and P-tRNA | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8YOO
 
 | | Cryo-EM structure of the human 80S ribosome with 100 um Tigecycline | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Denk, T, Cheng, J. | | Deposit date: | 2024-03-13 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8YOP
 
 | | Cryo-EM structure of the human 80S ribosome with 4 um Tigecycline | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Denk, T, Cheng, J. | | Deposit date: | 2024-03-13 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8XSY
 
 | | Cryo-EM structure of the human 80S ribosome with Tigecycline, e-tRNA and CCDC124 (40S head Swivelled) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8XT2
 
 | | Cryo-EM structure of the human 55S mitoribosome with 10uM Tigecycline | | Descriptor: | 12s rRNA, 16s rRNA, 39S ribosomal protein L22, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8XT1
 
 | | Cryo-EM structure of the human 39S mitoribosome with 5uM Tigecycline | | Descriptor: | 16s rRNA, 39S ribosomal protein L22, mitochondrial, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
6EDQ
 
 | | Crystal Structure of the Light-Gated Anion Channelrhodopsin GtACR1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Anion channelrhodopsin 1, GLYCEROL | | Authors: | Li, H, Huang, C.Y, Wang, M, Zheng, L, Spudich, J.L. | | Deposit date: | 2018-08-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a natural light-gated anion channelrhodopsin.
Elife, 8, 2019
|
|
8Q2O
 
 | | Structure of alginate transporter AlgE from P. aeruginosa PAO1 by using Se-MAG for the the lipid cubic phase crystallization | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Alginate production protein AlgE, ... | | Authors: | Huang, C.-Y, Boland, C, Kaki, S.S, Wang, M, Olieric, V, Caffrey, M. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Se-MAG Is a Convenient Additive for Experimental Phasing and Structure Determination of Membrane Proteins Crystallised by the Lipid Cubic Phase (In Meso) Method
Crystals, 2023
|
|
8Q2P
 
 | | Structure of the membrane integral lipoprotein N-acyltransferase Lnt from E. coli by using Se-MAG for the the lipid cubic phase crystallization | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Apolipoprotein N-acyltransferase, ... | | Authors: | Huang, C.-Y, Boland, C, Kaki, S.S, Wang, M, Olieric, V, Caffrey, M. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Se-MAG Is a Convenient Additive for Experimental Phasing and Structure Determination of Membrane Proteins Crystallised by the Lipid Cubic Phase (In Meso) Method
Crystals, 2023
|
|
2D3U
 
 | | X-ray crystal structure of hepatitis C virus RNA dependent RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | 5-(4-CYANOPHENYL)-3-{[(2-METHYLPHENYL)SULFONYL]AMINO}THIOPHENE-2-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-02 | | Release date: | 2006-08-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
2DHS
 
 | | Solution Structure of Nucleic Acid Binding Protein CUGBP1ab and its Binding Study with DNA and RNA | | Descriptor: | CUG triplet repeat RNA-binding protein 1 | | Authors: | Xia, Y.L, Jun, K.Y, Zhu, Q, Han, X.G, Zhang, H, Timchenko, L, Swanson, M, Gao, X.L. | | Deposit date: | 2006-03-25 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Nucleic Acid Binding Protein CUGBP1ab and its Binding Study with DNA and RNA
To be Published
|
|
5C3I
 
 | | Crystal structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2 | | Descriptor: | DNA replication licensing factor MCM2,MCM2, Histone H3.1, Histone H4, ... | | Authors: | Wang, H, Wang, M, Yang, N, Xu, R.M. | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2
Protein Cell, 6, 2015
|
|
6KHX
 
 | | Crystal structure of Prx from Akkermansia muciniphila | | Descriptor: | CALCIUM ION, Peroxiredoxin | | Authors: | Li, M, Wang, J, Xu, W, Wang, Y, Zhang, M, Wang, M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of Akkermansia muciniphila peroxiredoxin reveals a novel regulatory mechanism of typical 2-Cys Prxs by a distinct loop.
Febs Lett., 594, 2020
|
|
8QI8
 
 | | Cryogenic temperature dark state structure of CrPhotLOV1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-11 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
8QI9
 
 | | CrPhotLOV1 dark state structure determined by serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-11 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
8QIH
 
 | | CrPhotLOV1 light state structure 22.5 ms (20-25 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
8QIL
 
 | | CrPhotLOV1 light state structure 37.5 ms (35-40 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
8QIN
 
 | | CrPhotLOV1 light state structure 47.5 ms (45-50 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
