7KF1
 
 | |
7KF0
 
 | |
3UCS
 
 | |
3UOX
 
 | | Crystal Structure of OTEMO (FAD bound form 2) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, OTEMO | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UOY
 
 | | Crystal Structure of OTEMO complex with FAD and NADP (form 1) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO, ... | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UOV
 
 | | Crystal Structure of OTEMO (FAD bound form 1) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, OTEMO | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.045 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UP5
 
 | | Crystal Structure of OTEMO complex with FAD and NADP (form 4) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.453 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UP4
 
 | | Crystal Structure of OTEMO complex with FAD and NADP (form 3) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UOZ
 
 | | Crystal Structure of OTEMO complex with FAD and NADP (form 2) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
6IE8
 
 | | RamR in complex with cholic acid | | Descriptor: | CHOLIC ACID, Regulatory protein, SULFATE ION | | Authors: | Nakashima, R, Sakurai, K, Yamasaki, S, Nishino, K. | | Deposit date: | 2018-09-13 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the multidrug resistance regulator RamR complexed with bile acids.
Sci Rep, 9, 2019
|
|
6J1C
 
 | | Photoswitchable fluorescent protein Gamillus, N150C/T204V double mutant, off-state | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein | | Authors: | Nakashima, R, Shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
6J1B
 
 | | Photoswitchable fluorescent protein Gamillus, N150C/T204V double mutant, on-state | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Nakashima, R, Shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
5B83
 
 | |
8JQQ
 
 | | Protocatecuate hydroxylase from Xylophilus ampelinus C347T mutant | | Descriptor: | 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fukushima, R, Katsuki, N, Fushinobu, S, Takaya, N. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Protocatechuate hydroxylase is a novel group A flavoprotein monooxygenase with a unique substrate recognition mechanism.
J.Biol.Chem., 300, 2023
|
|
8JQP
 
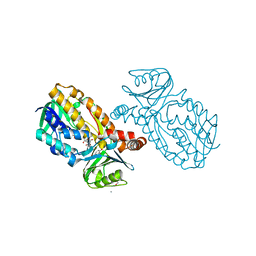 | | Protocatecuate hydroxylase from Xylophilus ampelinus complexed with 3,4-dihydroxybenzoate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), CALCIUM ION, ... | | Authors: | Fukushima, R, Katsuki, N, Fushinobu, S, Takaya, N. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Protocatechuate hydroxylase is a novel group A flavoprotein monooxygenase with a unique substrate recognition mechanism.
J.Biol.Chem., 300, 2023
|
|
8JQO
 
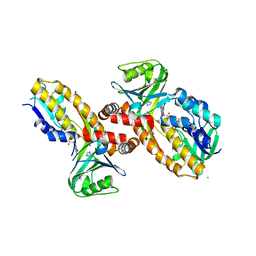 | | Protocatecuate hydroxylase from Xylophilus ampelinus complexed with imidazole | | Descriptor: | 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Fukushima, R, Katsuki, N, Fushinobu, S, Takaya, N. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protocatechuate hydroxylase is a novel group A flavoprotein monooxygenase with a unique substrate recognition mechanism.
J.Biol.Chem., 300, 2023
|
|
5I20
 
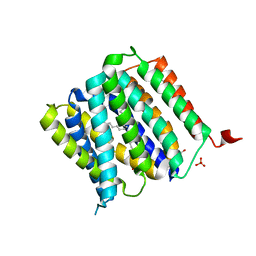 | | Crystal structure of protein | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, SULFATE ION, Uncharacterized protein | | Authors: | Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for amino acid export by DMT superfamily transporter YddG.
Nature, 534, 2016
|
|
6IE9
 
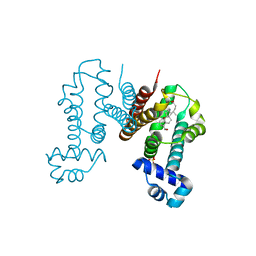 | | RamR in complex with chenodeoxycholic acid | | Descriptor: | CHENODEOXYCHOLIC ACID, Regulatory protein, SULFATE ION | | Authors: | Nakashima, R, Sakurai, K, Yamasaki, S, Nishino, K. | | Deposit date: | 2018-09-13 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the multidrug resistance regulator RamR complexed with bile acids.
Sci Rep, 9, 2019
|
|
6ZBT
 
 | | Structure of 14-3-3 gamma in complex with Nedd4-2 14-3-3 binding motif Ser342 | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, 14-3-3 protein gamma, E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Joshi, R, Kalabova, D, Obsil, T, Obsilova, V. | | Deposit date: | 2020-06-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79948521 Å) | | Cite: | 14-3-3-protein regulates Nedd4-2 by modulating interactions between HECT and WW domains.
Commun Biol, 4, 2021
|
|
6IIA
 
 | | MexB in complex with LMNG | | Descriptor: | Lauryl Maltose Neopentyl Glycol, Multidrug resistance protein MexB | | Authors: | Nakashima, R, Sakurai, K, Nakao, K. | | Deposit date: | 2018-10-04 | | Release date: | 2019-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structures of multidrug efflux pump MexB bound with high-molecular-mass compounds.
Sci Rep, 9, 2019
|
|
3AY5
 
 | |
8JMR
 
 | | Crystal structure of hinokiresinol synthase in complex with 1,7-bis(4-hydroxyphenyl)hepta-1,6-dien-3-one | | Descriptor: | 1,7-bis(4-hydroxyphenyl)hepta-1,6-dien-3-one, Hinokiresinol synthase alpha subunit, Hinokiresinol synthase beta subunit, ... | | Authors: | Ding, Y, Ushimaru, R, Mori, T, Abe, I. | | Deposit date: | 2023-06-05 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Mechanistic Insights into the C-C Bond-Forming Rearrangement Reaction Catalyzed by Heterodimeric Hinokiresinol Synthase.
J.Am.Chem.Soc., 145, 2023
|
|
8JMQ
 
 | | Crystal structure of hinokiresinol synthase | | Descriptor: | Hinokiresinol synthase alpha subunit, Hinokiresinol synthase beta subunit | | Authors: | Ding, Y, Ushimaru, R, Mori, T, Abe, I. | | Deposit date: | 2023-06-05 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insights into the C-C Bond-Forming Rearrangement Reaction Catalyzed by Heterodimeric Hinokiresinol Synthase.
J.Am.Chem.Soc., 145, 2023
|
|
8IAX
 
 | | Crystal structure of Streptococcus pneumoniae pyruvate kinase in complex with phosphoenolpyruvate and fructose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Nakashima, R, Taguchi, A. | | Deposit date: | 2023-02-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional and structural characterization of Streptococcus pneumoniae pyruvate kinase involved in fosfomycin resistance.
J.Biol.Chem., 299, 2023
|
|
8IAT
 
 | |
