6E5D
 
 | |
1FKV
 
 | | RECOMBINANT GOAT ALPHA-LACTALBUMIN T29I | | Descriptor: | ALPHA-LACTALBUMIN, CALCIUM ION | | Authors: | Horii, K, Matsushima, M, Tsumoto, K, Kumagai, I. | | Deposit date: | 2000-08-10 | | Release date: | 2001-02-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of Thr29 to the thermodynamic stability of goat alpha-lactalbumin as determined by experimental and theoretical approaches.
Proteins, 45, 2001
|
|
1FKQ
 
 | | RECOMBINANT GOAT ALPHA-LACTALBUMIN T29V | | Descriptor: | ALPHA-LACTALBUMIN, CALCIUM ION | | Authors: | Horii, K, Matsushima, M, Tsumoto, K, Kumagai, I. | | Deposit date: | 2000-08-10 | | Release date: | 2001-02-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of Thr29 to the thermodynamic stability of goat alpha-lactalbumin as determined by experimental and theoretical approaches.
Proteins, 45, 2001
|
|
3A4F
 
 | | Crystal Structure of Human Transthyretin (E54K) | | Descriptor: | GLYCEROL, SULFATE ION, Transthyretin | | Authors: | Miyata, M, Sato, T, Nakamura, T, Ikemizu, S, Yamagata, Y, Kai, H. | | Deposit date: | 2009-07-06 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Role of the glutamic acid 54 residue in transthyretin stability and thyroxine binding
Biochemistry, 49, 2010
|
|
3A4E
 
 | | Crystal structure of Human Transthyretin (E54G) | | Descriptor: | GLYCEROL, SULFATE ION, Transthyretin | | Authors: | Miyata, M, Sato, T, Nakamura, T, Ikemizu, S, Yamagata, Y, Kai, H. | | Deposit date: | 2009-07-06 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Role of the glutamic acid 54 residue in transthyretin stability and thyroxine binding
Biochemistry, 49, 2010
|
|
3A4D
 
 | | Crystal structure of Human Transthyretin (wild-type) | | Descriptor: | GLYCEROL, SULFATE ION, Transthyretin | | Authors: | Miyata, M. | | Deposit date: | 2009-07-06 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of the Glutamic Acid 54 Residue in Transthyretin Stability and Thyroxine Binding
Biochemistry, 2009
|
|
6MNA
 
 | |
7WUX
 
 | | Crystal structure of AziU3/U2 complexed with (5S,6S)-O7-sulfo DADH from Streptomyces sahachiroi | | Descriptor: | (2S,5S,6S)-2,6-bis(azanyl)-5-oxidanyl-7-sulfooxy-heptanoic acid, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, AziU2, ... | | Authors: | Kurosawa, S, Yoshida, A, Tomita, T, Nishiyama, M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Enzymatic Aziridine Formation via Sulfate Elimination.
J.Am.Chem.Soc., 144, 2022
|
|
7WUW
 
 | | Crystal structure of AziU3/U2 from Streptomyces sahachiroi | | Descriptor: | AziU2, AziU3, MAGNESIUM ION, ... | | Authors: | Kurosawa, S, Yoshida, A, Tomita, T, Nishiyama, M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular Basis for Enzymatic Aziridine Formation via Sulfate Elimination.
J.Am.Chem.Soc., 144, 2022
|
|
3WOR
 
 | | Crystal structure of the DAP BII octapeptide complex | | Descriptor: | Angiotensin II, GLYCEROL, ZINC ION, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOQ
 
 | | Crystal structure of the DAP BII hexapeptide complex III | | Descriptor: | Angiotensin IV, GLYCEROL, ZINC ION, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WON
 
 | | Crystal structure of the DAP BII dipeptide complex III | | Descriptor: | GLYCEROL, TYROSINE, VALINE, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOL
 
 | | Crystal structure of the DAP BII dipeptide complex I | | Descriptor: | GLYCEROL, TYROSINE, VALINE, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOM
 
 | | Crystal structure of the DAP BII dipeptide complex II | | Descriptor: | GLYCEROL, TYROSINE, VALINE, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
7EF9
 
 | | Crystal structure of mouse MUTYH in complex with DNA containing AP site analogue:8-oxoG (Form II) | | Descriptor: | Adenine DNA glycosylase, DNA (5'-D(*AP*TP*GP*AP*GP*AP*CP*(8OG)P*GP*GP*GP*AP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*TP*CP*CP*CP*(3DR)P*GP*TP*CP*TP*C)-3'), ... | | Authors: | Nakamura, T, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2021-03-21 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of the mammalian adenine DNA glycosylase MUTYH: insights into the base excision repair pathway and cancer.
Nucleic Acids Res., 49, 2021
|
|
7EF8
 
 | | Crystal structure of mouse MUTYH in complex with DNA containing AP site analogue:8-oxoG (Form I) | | Descriptor: | Adenine DNA glycosylase, DNA (5'-D(*TP*AP*GP*TP*CP*CP*CP*(3DR)P*GP*TP*CP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*AP*CP*(8OG)P*GP*GP*GP*AP*CP*T)-3'), ... | | Authors: | Nakamura, T, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2021-03-21 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the mammalian adenine DNA glycosylase MUTYH: insights into the base excision repair pathway and cancer.
Nucleic Acids Res., 49, 2021
|
|
7EFA
 
 | |
3WOP
 
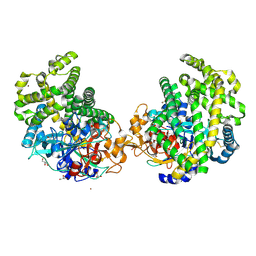 | | Crystal structure of the DAP BII hexapeptide complex II | | Descriptor: | Angiotensin IV, GLYCEROL, ZINC ION, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOI
 
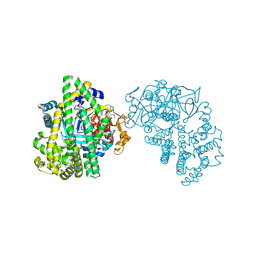 | | Crystal structure of the DAP BII (S657A) | | Descriptor: | GLYCEROL, ZINC ION, dipeptidyl aminopeptidase BII | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOO
 
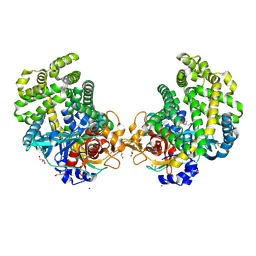 | | Crystal structure of the DAP BII hexapeptide complex I | | Descriptor: | Angiotensin II, GLYCEROL, ZINC ION, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOK
 
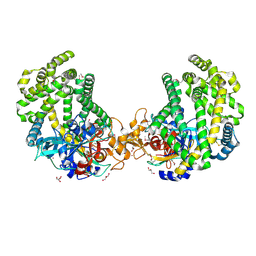 | | Crystal structure of the DAP BII (Space) | | Descriptor: | GLYCEROL, ZINC ION, dipeptidyl aminopeptidase BII | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOJ
 
 | | Crystal structure of the DAP BII | | Descriptor: | GLYCEROL, ZINC ION, dipeptidyl aminopeptidase BII | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
8IJU
 
 | | ATP-dependent RNA helicase DDX39A (URH49delta41) | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent RNA helicase DDX39A, PHOSPHATE ION, ... | | Authors: | Mikami, B, Fujita, K, Masuda, S, Kojima, M. | | Deposit date: | 2023-02-28 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural differences between the closely related RNA helicases, UAP56 and URH49, fashion distinct functional apo-complexes.
Nat Commun, 15, 2024
|
|
5X9P
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 5 | | Descriptor: | 3-[[4-chloranyl-2-nitro-5-[(2-oxidanylidene-1,3-dihydrobenzimidazol-5-yl)amino]phenyl]amino]propanoic acid, B-cell lymphoma 6 protein | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of a novel B-cell lymphoma 6 (BCL6)-corepressor interaction inhibitor by utilizing structure-based drug design
Bioorg. Med. Chem., 25, 2017
|
|
5X9O
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 1a | | Descriptor: | 1,2-ETHANEDIOL, 5-[(2-chloranyl-4-nitro-phenyl)amino]-1,3-dihydrobenzimidazol-2-one, B-cell lymphoma 6 protein, ... | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of a novel B-cell lymphoma 6 (BCL6)-corepressor interaction inhibitor by utilizing structure-based drug design
Bioorg. Med. Chem., 25, 2017
|
|
