5YL1
 
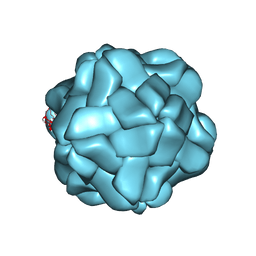 | | T=1 subviral particle of Penaeus vannamei nodavirus capsid protein deletion mutant (delta 1-37 & 251-368) | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
5XMJ
 
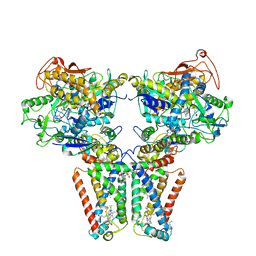 | | Crystal structure of quinol:fumarate reductase from Desulfovibrio gigas | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Guan, H.H, Hsieh, Y.C, Lin, P.R, Chen, C.J. | | Deposit date: | 2017-05-15 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insights into the electron/proton transfer pathways in the quinol:fumarate reductase from Desulfovibrio gigas.
Sci Rep, 8, 2018
|
|
6AB5
 
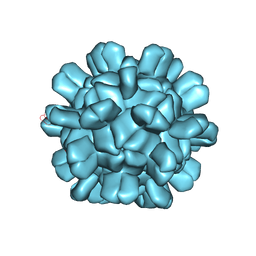 | | Cryo-EM structure of T=1 Penaeus vannamei nodavirus | | Descriptor: | Capsid protein | | Authors: | Chen, N.C, Miyazaki, N, Yoshimura, M, Guan, H.H, Lin, C.C, Iwasaki, K, Chen, C.J. | | Deposit date: | 2018-07-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
6AB6
 
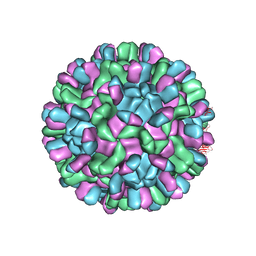 | | Cryo-EM structure of T=3 Penaeus vannamei nodavirus | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Chen, N.C, Miyazaki, N, Yoshimura, M, Guan, H.H, Lin, C.C, Iwasaki, K, Chen, C.J. | | Deposit date: | 2018-07-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
8W4L
 
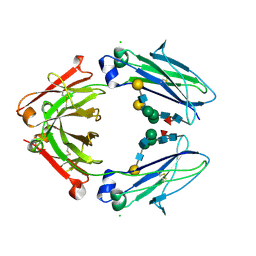 | | Crystal structure of closed conformation of human immunoglobulin Fc in presence of EndoSz | | Descriptor: | CHLORIDE ION, Immunoglobulin gamma-1 heavy chain, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guan, H.H, Lin, C.C, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based High-Efficiency Homogeneous Antibody Platform by Endoglycosidase Sz Provides Insights into Its Transglycosylation Mechanism.
Jacs Au, 4, 2024
|
|
8W4M
 
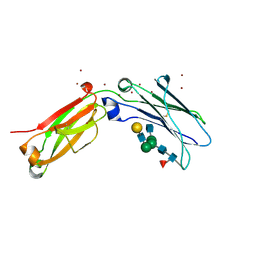 | | Crystal structure of open conformation of human immunoglobulin Fc in presence of EndoSz | | Descriptor: | Immunoglobulin gamma-1 heavy chain, ZINC ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Guan, H.H, Lin, C.C, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-Based High-Efficiency Homogeneous Antibody Platform by Endoglycosidase Sz Provides Insights into Its Transglycosylation Mechanism.
Jacs Au, 4, 2024
|
|
8W4G
 
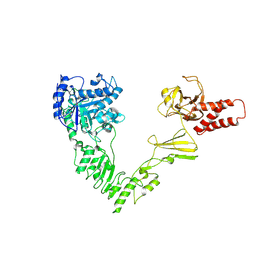 | | Crystal structure of EndoSz mutant D234M, from Streptococcus equi subsp. Zooepidemicus Sz105 | | Descriptor: | CALCIUM ION, glycoside hydrolase | | Authors: | Guan, H.H, Lin, C.C, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based High-Efficiency Homogeneous Antibody Platform by Endoglycosidase Sz Provides Insights into Its Transglycosylation Mechanism.
Jacs Au, 4, 2024
|
|
8W4N
 
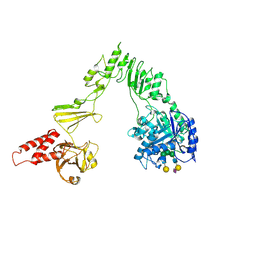 | | Crystal structure of EndoSz mutant D234M, in space group P21, in complex with oligosaccharide G2S1 | | Descriptor: | CALCIUM ION, Glycoside hydrolase, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Guan, H.H, Lin, C.C, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based High-Efficiency Homogeneous Antibody Platform by Endoglycosidase Sz Provides Insights into Its Transglycosylation Mechanism.
Jacs Au, 4, 2024
|
|
8W4I
 
 | | Crystal structure of EndoSz mutant D234M in space group P21 | | Descriptor: | CALCIUM ION, glycoside hydrolase | | Authors: | Guan, H.H, Lin, C.C, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Based High-Efficiency Homogeneous Antibody Platform by Endoglycosidase Sz Provides Insights into Its Transglycosylation Mechanism.
Jacs Au, 4, 2024
|
|
7B21
 
 | | The X183 domain from Cellvibrio japonicus Cbp2D | | Descriptor: | 1,2-ETHANEDIOL, Carbohydrate binding protein, putative, ... | | Authors: | Branch, J, Hemsworth, G.R. | | Deposit date: | 2020-11-25 | | Release date: | 2021-07-21 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | C-type cytochrome-initiated reduction of bacterial lytic polysaccharide monooxygenases.
Biochem.J., 478, 2021
|
|
7XPE
 
 | | Cryo-EM structure of the T=4 lake sinai virus 2 virus-like capsid at pH 8.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-05-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions.
Nat Commun, 14, 2023
|
|
7XGZ
 
 | | Cryo-EM structure of the T=4 lake sinai virus 2 virus-like capsid at pH 7.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-04-07 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions
Nat Commun, 14, 2023
|
|
7XPA
 
 | | Cryo-EM structure of the T=3 lake sinai virus 2 virus-like capsid at pH 7.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-05-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions.
Nat Commun, 14, 2023
|
|
7XPD
 
 | | Cryo-EM structure of the T=3 lake sinai virus 2 virus-like capsid at pH 6.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-05-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions.
Nat Commun, 14, 2023
|
|
7XPF
 
 | | Cryo-EM structure of the T=3 lake sinai virus 2 virus-like capsid at pH 8.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-05-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions.
Nat Commun, 14, 2023
|
|
7XPB
 
 | | Cryo-EM structure of the T=4 lake sinai virus 2 virus-like capsid at pH 6.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-05-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions.
Nat Commun, 14, 2023
|
|
7XPG
 
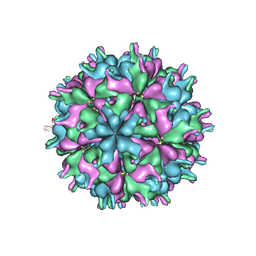 | | Cryo-EM structure of the T=3 lake sinai virus 1 (delta-N48) virus-like capsid at pH 6.5 | | Descriptor: | Capsid protein alpha, RNA (5'-R(P*UP*G)-3') | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-05-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions.
Nat Commun, 14, 2023
|
|
6Z5Y
 
 | |
8GPB
 
 | |
6I0Y
 
 | | TnaC-stalled ribosome complex with the titin I27 domain folding close to the ribosomal exit tunnel | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Su, T, Kudva, R, von Heijne, G, Beckmann, R. | | Deposit date: | 2018-10-26 | | Release date: | 2018-12-05 | | Last modified: | 2019-01-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Folding pathway of an Ig domain is conserved on and off the ribosome.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8B5Q
 
 | |
8B5S
 
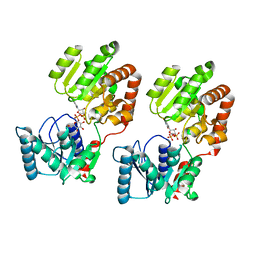 | | Crystal Structure of P. aeruginosa WaaG in complex with UDP-glucose | | Descriptor: | UDP-glucose:(Heptosyl) LPS alpha 1,3-glucosyltransferase WaaG, URIDINE-5'-DIPHOSPHATE, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Scaletti, E, Gustafsson Westergren, R, Stenmark, P. | | Deposit date: | 2022-09-24 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and functional insights into the Pseudomonas aeruginosa glycosyltransferase WaaG and the implications for lipopolysaccharide biosynthesis.
J.Biol.Chem., 299, 2023
|
|
8B62
 
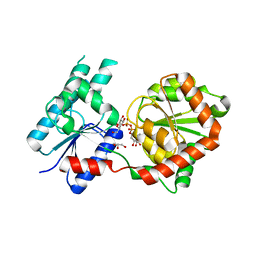 | | Crystal Structure of P. aeruginosa WaaG in complex with UDP-galactose | | Descriptor: | GALACTOSE-URIDINE-5'-DIPHOSPHATE, GLYCEROL, UDP-glucose:(Heptosyl) LPS alpha 1,3-glucosyltransferase WaaG | | Authors: | Scaletti, E, Gustafsson Westergren, R, Stenmark, P. | | Deposit date: | 2022-09-25 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural and functional insights into the Pseudomonas aeruginosa glycosyltransferase WaaG and the implications for lipopolysaccharide biosynthesis.
J.Biol.Chem., 299, 2023
|
|
8B63
 
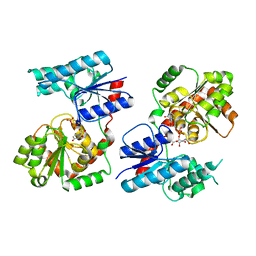 | | Crystal Structure of P. aeruginosa WaaG in complex with UDP-GalNAc | | Descriptor: | ACETATE ION, UDP-glucose:(Heptosyl) LPS alpha 1,3-glucosyltransferase WaaG, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Scaletti, E, Gustafsson Westergren, R, Stenmark, P. | | Deposit date: | 2022-09-25 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insights into the Pseudomonas aeruginosa glycosyltransferase WaaG and the implications for lipopolysaccharide biosynthesis.
J.Biol.Chem., 299, 2023
|
|
6GPB
 
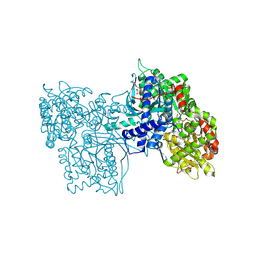 | |
