5TPK
 
 | |
3M0J
 
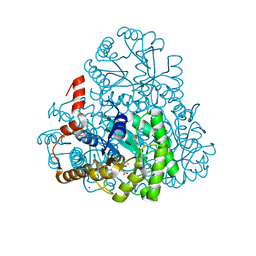 | | Structure of oxaloacetate acetylhydrolase in complex with the inhibitor 3,3-difluorooxalacetate | | Descriptor: | 2,2-difluoro-3,3-dihydroxybutanedioic acid, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Herzberg, O, Chen, C. | | Deposit date: | 2010-03-03 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of oxalacetate acetylhydrolase, a virulence factor of the chestnut blight fungus.
J.Biol.Chem., 285, 2010
|
|
3M0K
 
 | |
3LYE
 
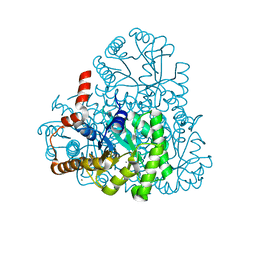 | | Crystal structure of oxaloacetate acetylhydrolase | | Descriptor: | CALCIUM ION, Oxaloacetate acetyl hydrolase | | Authors: | Herzberg, O, Chen, C. | | Deposit date: | 2010-02-26 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of oxalacetate acetylhydrolase, a virulence factor of the chestnut blight fungus.
J.Biol.Chem., 285, 2010
|
|
6A4C
 
 | | Solution structure of MXAN_0049 | | Descriptor: | Uncharacterized protein MXAN_0049 | | Authors: | Chen, C, Feng, Y. | | Deposit date: | 2018-06-19 | | Release date: | 2019-06-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of MXAN_0049
To Be Published
|
|
3QYM
 
 | |
7FCF
 
 | | Crystal structure of T6SS Hcp protein | | Descriptor: | Fimbrial protein | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Bacterial antagonism of Chromobacterium haemolyticum and characterization of its putative type VI secretion system.
Res.Microbiol., 173, 2022
|
|
5XWR
 
 | | Crystal Structure of RBBP4-peptide complex | | Descriptor: | Histone-binding protein RBBP4, MET-SER-ARG-ARG-LYS-GLN-ALA-LYS-PRO-GLN-HIS-ILE | | Authors: | Jobichen, C, Lui, B.H, Daniel, G.T, Sivaraman, J. | | Deposit date: | 2017-06-30 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Targeting cancer addiction for SALL4 by shifting its transcriptome with a pharmacologic peptide.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4PLQ
 
 | |
4ZQY
 
 | |
3TVD
 
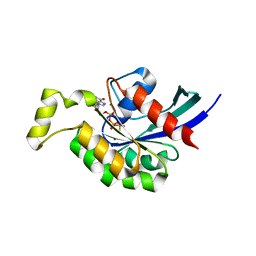 | | Crystal Structure of Mouse RhoA-GTP complex | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Swaminathan, K, Pal, K, Jobichen, C. | | Deposit date: | 2011-09-20 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Crystal structure of mouse RhoA:GTPgammaS complex in a centered lattice.
J.Struct.Funct.Genom., 13, 2012
|
|
3VTS
 
 | |
4RUD
 
 | | Crystal structure of a three finger toxin | | Descriptor: | Three-finger toxin 3b, ZINC ION | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2014-11-18 | | Release date: | 2016-05-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fulditoxin, representing a new class of dimeric snake toxins, defines novel pharmacology at nicotinic ACh receptors.
Br.J.Pharmacol., 177, 2020
|
|
7DEY
 
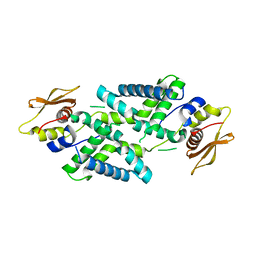 | | Structure of Dicer from Pichia stipitis | | Descriptor: | RNase III | | Authors: | Jobichen, C, Jingru, C. | | Deposit date: | 2020-11-05 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | Structural and mechanistic insight into stem-loop RNA processing by yeast Pichia stipitis Dicer.
Protein Sci., 30, 2021
|
|
7C28
 
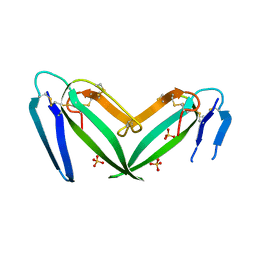 | | Unusual quaternary structure of a homodimeric synergistic toxin from mamba snake venom | | Descriptor: | SULFATE ION, Synergistic-type venom protein S2C4 | | Authors: | Jobichen, C, Narumi, A, Sivaraman, J, Kini, R.M. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Unusual quaternary structure of a homodimeric synergistic-type toxin from mamba snake venom defines its molecular evolution.
Biochem.J., 477, 2020
|
|
3D23
 
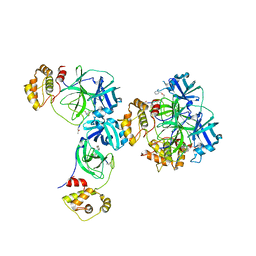 | | Main protease of HCoV-HKU1 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Zhao, Q, Chen, C, Li, S, Zou, Y. | | Deposit date: | 2008-05-07 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the main protease from a global infectious human coronavirus, HCoV-HKU1.
J.Virol., 82, 2008
|
|
4ZXU
 
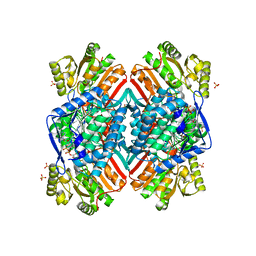 | | 2.85 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) H448F/P449M double mutant from Staphylococcus aureus in complex with NAD+ and BME-free Cys289 | | Descriptor: | Betaine-aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | 2.85 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) H448F/P449M double mutant from Staphylococcus aureus in complex with NAD+ and BME-free Cys289.
To be Published
|
|
5DIB
 
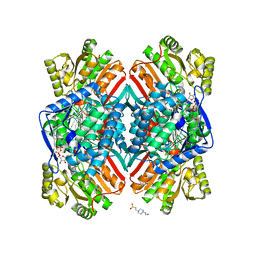 | | 2.25 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) Y450L point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-08-31 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2.25 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) Y450L point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289
To Be Published
|
|
5EZ4
 
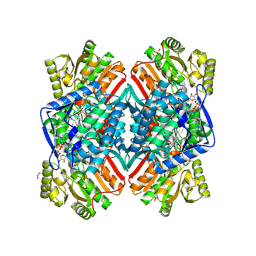 | | 2.11 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M/Y450L double mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-26 | | Release date: | 2015-12-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | 2.11 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M/Y450L double mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289
To Be Published
|
|
5EYU
 
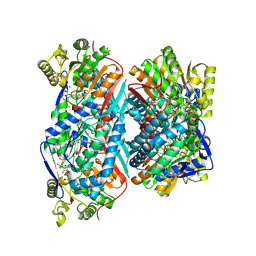 | | 1.72 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-25 | | Release date: | 2015-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | 1.72 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289
To Be Published
|
|
5Y6N
 
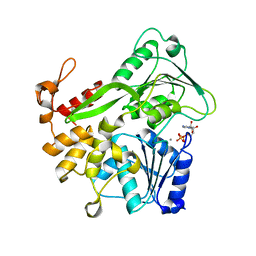 | | Zika virus helicase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Helicase domain from Genome polyprotein, MANGANESE (II) ION | | Authors: | Yang, X.Y, Chen, C, Tian, H.L, Chi, H, Mu, Z.Y, Zhang, T.Q, Yang, K.L, Zhao, Q, Liu, X.H, Wang, Z.F, Ji, X.Y, Yang, H.T. | | Deposit date: | 2017-08-12 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Mechanism of ATP hydrolysis by the Zika virus helicase.
FASEB J., 32, 2018
|
|
4MY2
 
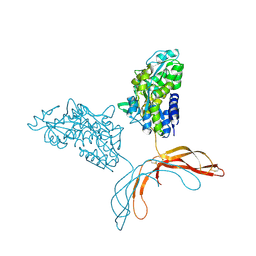 | | Crystal Structure of Norrin in fusion with Maltose Binding Protein | | Descriptor: | Maltose-binding periplasmic protein, Norrin fusion protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ke, J, Jurecky, C, Chen, C, Gu, X, Parker, N, Williams, B.O, Melcher, K, Xu, H.E. | | Deposit date: | 2013-09-27 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and function of Norrin in assembly and activation of a Frizzled 4-Lrp5/6 complex.
Genes Dev., 27, 2013
|
|
1KVZ
 
 | | Solution Structure of Cytotoxic RC-RNase4 | | Descriptor: | RC-RNase4 | | Authors: | Hsu, C.-H, Liao, Y.-D, Chen, L.-W, Wu, S.-H, Chen, C. | | Deposit date: | 2002-01-28 | | Release date: | 2002-07-28 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Cytotoxic RNase 4 from the Oocytes of Bullfrog Rana Catesbeiana
J.MOL.BIOL., 326, 2003
|
|
5X3L
 
 | |
1BC4
 
 | | THE SOLUTION STRUCTURE OF A CYTOTOXIC RIBONUCLEASE FROM THE OOCYTES OF RANA CATESBEIANA (BULLFROG), NMR, 15 STRUCTURES | | Descriptor: | RIBONUCLEASE | | Authors: | Chang, C.-F, Chen, C, Chen, Y.-C, Hom, K, Huang, R.-F, Huang, T. | | Deposit date: | 1998-05-05 | | Release date: | 1998-10-14 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a cytotoxic ribonuclease from the oocytes of Rana catesbeiana (bullfrog).
J.Mol.Biol., 283, 1998
|
|
