6AKI
 
 | | Calcium release-activated calcium channel protein 1, P288L mutant | | Descriptor: | CHLORIDE ION, Calcium release-activated calcium channel protein 1 | | Authors: | Liu, X, Wu, G, Yang, X, Shen, Y. | | Deposit date: | 2018-09-01 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (4.496 Å) | | Cite: | Calcium release-activated calcium channel protein 1, P288L mutant
To Be Published
|
|
6K81
 
 | | Crystal structure of human VASH1-SVBP complex | | Descriptor: | Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | Authors: | Liu, X, Wang, H, Zhang, Y, Feng, Y. | | Deposit date: | 2019-06-11 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural insights into tubulin detyrosination by vasohibins-SVBP complex.
Cell Discov, 5, 2019
|
|
8GUB
 
 | | Cryo-EM structure of cancer-specific PI3Kalpha mutant H1047R in complex with BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Zhou, Q, Hart, J.R, Xu, Y, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-09-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8GUD
 
 | | Cryo-EM structure of cancer-specific PI3Kalpha mutant E545K in complex with BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Zhou, Q, Hart, J.R, Xu, Y, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-09-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8GUA
 
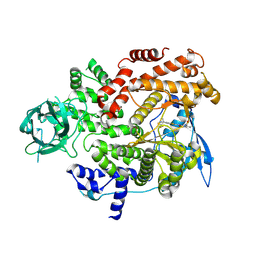 | | Cryo-EM structure of cancer-specific PI3Kalpha mutant E542K in complex with BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Zhou, Q, Hart, J.R, Xu, Y, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-09-11 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5ZNU
 
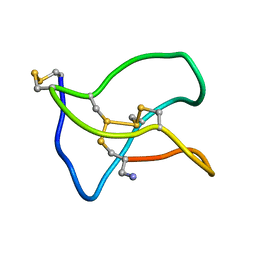 | | Structure of omega conotoxin Bu8 | | Descriptor: | Omega-conotoxin-like Bu8 | | Authors: | Liu, X, Jiang, L. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-17 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of omega conotoxin bu8
To Be Published
|
|
4UEI
 
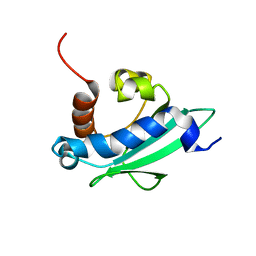 | | Solution structure of the sterol carrier protein domain 2 of Helicoverpa armigera | | Descriptor: | STEROL CARRIER PROTEIN 2/3-OXOACYL-COA THIOLASE | | Authors: | Liu, X, Ma, H, Yan, X, Hong, H, Peng, J, Peng, R. | | Deposit date: | 2014-12-18 | | Release date: | 2015-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Function of Helicoverpa Armigera Sterol Carrier Protein-2, an Important Insecticidal Target from the Cotton Bollworm.
Sci.Rep., 5, 2015
|
|
6EG8
 
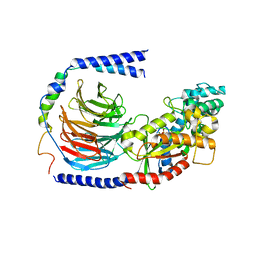 | | Structure of the GDP-bound Gs heterotrimer | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Hilger, D, Liu, X, Aschauer, P, Kobilka, B.K. | | Deposit date: | 2018-08-19 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Process of GPCR-G Protein Complex Formation.
Cell, 177, 2019
|
|
6LU7
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor N3 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | Deposit date: | 2020-01-26 | | Release date: | 2020-02-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
8I6Y
 
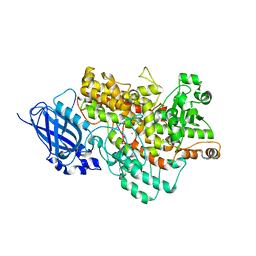 | | Crystal structure of Arabidopsis thaliana LOX1 | | Descriptor: | FE (III) ION, Linoleate 9S-lipoxygenase 1 | | Authors: | Liu, X, Liu, L. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | UV-B light signal mediates stomatal closure by activating the 9-lipoxygenase pathway
To Be Published
|
|
3X1D
 
 | |
3QOF
 
 | |
3QNU
 
 | |
6KSB
 
 | |
6LQ3
 
 | |
6LQ7
 
 | |
6LPY
 
 | |
6LQ5
 
 | |
5XMZ
 
 | | Verticillium effector PevD1 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Effector protein PevD1 | | Authors: | Liu, X, Zhou, R. | | Deposit date: | 2017-05-17 | | Release date: | 2017-07-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The asparagine-rich protein NRP interacts with the Verticillium effector PevD1 and regulates the subcellular localization of cryptochrome 2
J. Exp. Bot., 68, 2017
|
|
5GQQ
 
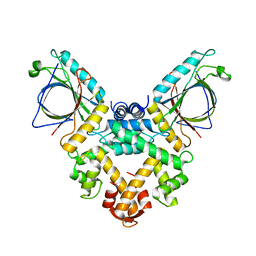 | | Structure of ALG-2/HEBP2 Complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Heme-binding protein 2, ... | | Authors: | Liu, X, Ma, J, Zhang, H, Feng, Y. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Study of Apoptosis-linked Gene-2Heme-binding Protein 2 Interactions in HIV-1 Production.
J. Biol. Chem., 291, 2016
|
|
3EXT
 
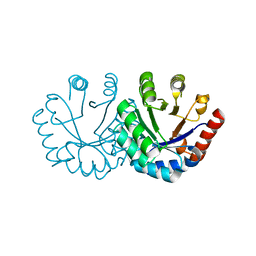 | | Crystal structure of KGPDC from Streptococcus mutans | | Descriptor: | MAGNESIUM ION, RmpD (Hexulose-6-phosphate synthase) | | Authors: | Liu, X, Li, G.L, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
6LQ8
 
 | |
3S2S
 
 | |
3S70
 
 | | Crystal structure of active caspase-6 bound with Ac-VEID-CHO solved by As-SAD | | Descriptor: | ACETATE ION, CACODYLATE ION, Caspase-6, ... | | Authors: | Su, X.-D, Liu, X, Wang, X.-J. | | Deposit date: | 2011-05-26 | | Release date: | 2012-04-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.625 Å) | | Cite: | Get phases from arsenic anomalous scattering: de novo SAD phasing of two protein structures crystallized in cacodylate buffer
Plos One, 6, 2011
|
|
7JXG
 
 | | Structural model for Fe-containing human acireductone dioxygenase | | Descriptor: | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase, FE (II) ION | | Authors: | Pochapsky, T.C, Liu, X, Deshpande, A, Ringe, D, Garber, A, Ryan, J. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Model for the Solution Structure of Human Fe(II)-Bound Acireductone Dioxygenase and Interactions with the Regulatory Domain of Matrix Metalloproteinase I (MMP-I).
Biochemistry, 59, 2020
|
|
