4BUE
 
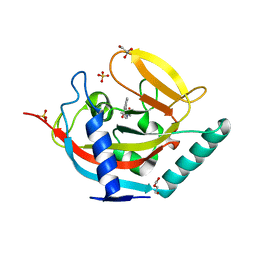 | | Crystal structure of human tankyrase 2 in complex with 3-methyl-N-(4-(4-oxo-3,4-dihydroquinazolin-2-yl)phenyl)butanamide | | Descriptor: | 3-METHYL-N-[4-(4-OXO-3,4-DIHYDROQUINAZOLIN-2-YL)PHENYL)BUTANAMIDE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Para-Substituted 2-Phenyl-3,4-Dihydroquinazolin-4-Ones as Potent and Selective Tankyrase Inhibitors.
Chemmedchem, 8, 2013
|
|
6FJ1
 
 | | Structure of the Ldtfm-avibactam carbamoyl enzyme | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 3,6,9,12,15-PENTAOXAHEPTADECANE, CHLORIDE ION, ... | | Authors: | Li de la Sierra Gallay, I, Iannazzo, L, Compain, F, Fonvielle, M, van Tilbeurgh, H, Edoo, Z, Arthur, M, Etheve-Quelquejeu, M, Hugonnet, J. | | Deposit date: | 2018-01-19 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Synthesis of Avibactam Derivatives and Activity on beta-Lactamases and Peptidoglycan Biosynthesis Enzymes of Mycobacteria.
Chemistry, 24, 2018
|
|
4AUQ
 
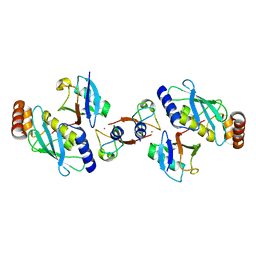 | | Structure of BIRC7-UbcH5b-Ub complex. | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 7, POLYUBIQUITIN-C, UBIQUITIN-CONJUGATING ENZYME E2 D2, ... | | Authors: | Dou, H, Buetow, L, Sibbet, G.J, Cameron, K, Huang, D.T. | | Deposit date: | 2012-05-21 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.176 Å) | | Cite: | Birc7-E2 Ubiquitin Conjugate Structure Reveals the Mechanism of Ubiquitin Transfer by a Ring Dimer.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1ORX
 
 | | Solution Structure of the acyclic permutant des-(24-28)-kalata B1. | | Descriptor: | kalata B1 | | Authors: | Barry, D.G, Daly, N.L, Clark, R.J, Sando, L, Craik, D.J. | | Deposit date: | 2003-03-17 | | Release date: | 2003-06-24 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Linearization of a naturally occurring circular protein maintains structure but eliminates hemolytic activity
Biochemistry, 42, 2003
|
|
1OSD
 
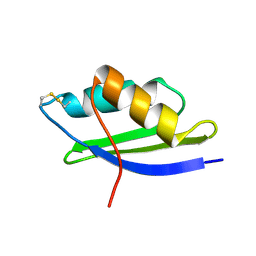 | | crystal structure of Oxidized MerP from Ralstonia metallidurans CH34 | | Descriptor: | hypothetical protein MerP | | Authors: | Serre, L, Rossy, E, Pebay-Peyroula, E, Cohen-Addad, C, Coves, J. | | Deposit date: | 2003-03-19 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Oxidized Form of the Periplasmic Mercury-binding Protein MerP from Ralstonia metallidurans CH34
J.MOL.BIOL., 339, 2004
|
|
5N6L
 
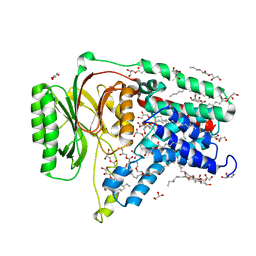 | | Structure of the membrane integral lipoprotein N-acyltransferase Lnt C387A mutant from E. coli | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, GLYCEROL | | Authors: | Huang, C.-Y, Boland, C, Howe, N, Wiktor, M, Vogeley, L, Weichert, D, Bailey, J, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis.
Nat Commun, 8, 2017
|
|
3SZN
 
 | |
6FQF
 
 | | THE X-RAY STRUCTURE OF FERRIC-BETA4 HUMAN HEMOGLOBIN | | Descriptor: | Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mazzarella, L, Merlino, A, Balasco, N, Balsamo, A, Vergara, A. | | Deposit date: | 2018-02-14 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the ferric homotetrameric beta4human hemoglobin.
Biophys. Chem., 240, 2018
|
|
1E20
 
 | | The FMN binding protein AtHal3 | | Descriptor: | BETA-MERCAPTOETHANOL, FLAVIN MONONUCLEOTIDE, HALOTOLERANCE PROTEIN HAL3, ... | | Authors: | Albert, A, Martinez-Ripoll, M, Espinosa-Ruiz, A, Yenush, L, Culianez-Macia, F.A, Serrano, R. | | Deposit date: | 2000-05-12 | | Release date: | 2000-09-11 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The X-Ray Structure of the Fmn-Binding Protein Athal3 Provides the Structural Basis for the Activity of a Regulatory Subunit Involved in Signal Transduction
Structure, 8, 2000
|
|
4B3P
 
 | | Structures of HIV-1 RT and RNA-DNA Complex Reveal a Unique RT Conformation and Substrate Interface | | Descriptor: | DNA, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H, ... | | Authors: | Lapkouski, M, Tian, L, Miller, J.T, Le Grice, S.F.J, Yang, W. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.839 Å) | | Cite: | Complexes of HIV-1 RT, Nnrti and RNA/DNA Hybrid Reveal a Structure Compatible with RNA Degradation
Nat.Struct.Mol.Biol., 20, 2013
|
|
4B2X
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-amino-1-butyl-5-(cyclopentylamino)pyrimidine-2,4(1H,3H)-dione, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Sarkar, A, Brenk, R, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-07-18 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
1OUV
 
 | | Helicobacter cysteine rich protein C (HcpC) | | Descriptor: | conserved hypothetical secreted protein | | Authors: | Mittl, P.R, Luethy, L. | | Deposit date: | 2003-03-25 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Helicobacter Cysteine-rich Protein C at 2.0A Resolution: Similar Peptide-binding Sites in TPR and SEL1-like Repeat Proteins
J.Mol.Biol., 340, 2004
|
|
5YH8
 
 | | The crystal structure of Staphylococcus aureus CntA in complex with staphylopine and nickel | | Descriptor: | (2~{S})-4-[[(2~{R})-3-(1~{H}-imidazol-4-yl)-1-oxidanyl-1-oxidanylidene-propan-2-yl]amino]-2-[[(2~{S})-1-oxidanyl-1-oxidanylidene-propan-2-yl]amino]butanoic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Ji, Q, Song, L. | | Deposit date: | 2017-09-27 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Mechanistic insights into staphylopine-mediated metal acquisition
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1E1S
 
 | | Human prion protein variant S170N | | Descriptor: | PRION PROTEIN | | Authors: | Calzolai, L, Lysek, D.A, Guntert, P, Von Schroetter, C, Zahn, R, Riek, R, Wuthrich, K. | | Deposit date: | 2000-05-10 | | Release date: | 2000-07-21 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Three Single-Residue Variants of the Human Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5YHE
 
 | | The crystal structure of Staphylococcus aureus CntA in complex with staphylopine and cobalt | | Descriptor: | (2~{S})-4-[[(2~{R})-3-(1~{H}-imidazol-4-yl)-1-oxidanyl-1-oxidanylidene-propan-2-yl]amino]-2-[[(2~{S})-1-oxidanyl-1-oxidanylidene-propan-2-yl]amino]butanoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Song, L, Ji, Q. | | Deposit date: | 2017-09-28 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.465 Å) | | Cite: | Mechanistic insights into staphylopine-mediated metal acquisition
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1OP4
 
 | | Solution Structure of Neural Cadherin Prodomain | | Descriptor: | Neural-cadherin | | Authors: | Koch, A.W, Farooq, A, Shan, W, Zeng, L, Colman, D.R, Zhou, M.-M. | | Deposit date: | 2003-03-04 | | Release date: | 2004-03-16 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure of the Neural (N-) Cadherin Prodomain Reveals a Cadherin Extracellular Domain-like Fold without Adhesive Characteristics
Structure, 12, 2004
|
|
1OYL
 
 | | Crystal Structures of the Ferric, Ferrous, and Ferrous-NO Forms of the Asp140Ala Mutant of Human Heme Oxygenase-1: Catalytic Implications | | Descriptor: | Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lad, L, Wang, J, Li, H, Friedman, J, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2003-04-05 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal structures of the ferric, ferrous, and ferrous-NO forms of the Asp140Ala mutant of human heme oxygenase-1: catalytic implications
J.Mol.Biol., 330, 2003
|
|
1OQV
 
 | |
1OYZ
 
 | | X-RAY STRUCTURE OF YIBA_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ET31. | | Descriptor: | Hypothetical protein yibA | | Authors: | Kuzin, A, Semesi, A, Skarina, T, Savchenko, A, Arrowsmith, C, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-04-07 | | Release date: | 2003-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-RAY STRUCTURE OF YIBA_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ET31.
To be Published
|
|
4ARV
 
 | | Yersinia kristensenii phytase apo form | | Descriptor: | 1,2-ETHANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, ... | | Authors: | Ariza, A, Moroz, O.V, Blagova, E.B, Turkenburg, J.P, Vevodova, J, Roberts, S, Vind, J, Sjoholm, C, Lassen, S.F, De Maria, L, Glitsoe, V, Skov, L.K, Wilson, K.S. | | Deposit date: | 2012-04-26 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Degradation of Phytate by the 6-Phytase from Hafnia Alvei: A Combined Structural and Solution Study.
Plos One, 8, 2013
|
|
4ASY
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Sarkar, A, Brenk, R, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-05-03 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
8UKC
 
 | | Solution NMR Structure of the lasso peptide chlorolassin | | Descriptor: | Lassopeptide Chlorolassin | | Authors: | Saad, H, Zhu, L, Harris, L.A, Shelton, K.E, Mitchell, D.A. | | Deposit date: | 2023-10-12 | | Release date: | 2024-09-25 | | Method: | SOLUTION NMR | | Cite: | Tryptophan-Centric Bioinformatics Identifies New Lasso Peptide Modifications.
Biochemistry, 63, 2024
|
|
5N5Q
 
 | | Human TTR altered conformation from soaking in iron chloride. | | Descriptor: | ACETATE ION, FE (III) ION, Transthyretin, ... | | Authors: | Ciccone, L, Savko, M, Shepard, W, Stura, E.A. | | Deposit date: | 2017-02-14 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Copper mediated amyloid-beta binding to Transthyretin.
Sci Rep, 8, 2018
|
|
8UKG
 
 | | Solution NMR structure of the lasso peptide wygwalassin-A1 | | Descriptor: | Lassopeptide wygwalassin-A1 | | Authors: | Saad, H, Zhu, L, Harris, L.A, Shelton, K.E, Mitchell, D.A. | | Deposit date: | 2023-10-13 | | Release date: | 2024-09-25 | | Method: | SOLUTION NMR | | Cite: | Tryptophan-Centric Bioinformatics Identifies New Lasso Peptide Modifications.
Biochemistry, 63, 2024
|
|
8RKB
 
 | |
