3UK1
 
 | |
3URR
 
 | |
3UW1
 
 | |
3V2I
 
 | |
3UW2
 
 | |
3V7C
 
 | | Cystal structure of SaBPL in complex with inhibitor | | Descriptor: | 5'-deoxy-2',3'-O-(1-methylethylidene)-5'-(4-{5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentyl}-1H-1,2,3-triazol-1-yl)adenosine, Biotin ligase | | Authors: | Yap, M.Y, Pendini, N.R. | | Deposit date: | 2011-12-20 | | Release date: | 2012-04-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Selective inhibition of biotin protein ligase from Staphylococcus aureus.
J.Biol.Chem., 287, 2012
|
|
3V8J
 
 | |
3V9O
 
 | |
2EU1
 
 | | Crystal structure of the chaperonin GroEL-E461K | | Descriptor: | GROEL | | Authors: | Cabo-Bilbao, A, Spinelli, S, Sot, B, Agirre, J, Mechaly, A.E, Muga, A, Guerin, D.M.A. | | Deposit date: | 2005-10-28 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Crystal structure of the temperature-sensitive and allosteric-defective chaperonin GroEL(E461K).
J.Struct.Biol., 155, 2006
|
|
2F2G
 
 | | X-Ray Structure of Gene Product From Arabidopsis Thaliana AT3G16990 | | Descriptor: | 4-AMINO-5-HYDROXYMETHYL-2-METHYLPYRIMIDINE, SEED MATURATION PROTEIN PM36 HOMOLOG, SULFATE ION | | Authors: | Wesenberg, G.W, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-11-16 | | Release date: | 2005-12-13 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of gene locus At3g16990 from Arabidopsis thaliana
Proteins, 57, 2004
|
|
1BGV
 
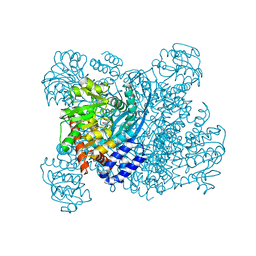 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE, GLUTAMIC ACID | | Authors: | Stillman, T.J, Baker, P.J, Britton, K.L, Rice, D.W. | | Deposit date: | 1998-06-01 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational flexibility in glutamate dehydrogenase. Role of water in substrate recognition and catalysis.
J.Mol.Biol., 234, 1993
|
|
8Y0Y
 
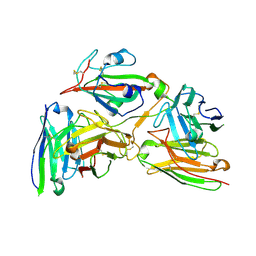 | | Cryo-EM structure of the 123-316 scDb/PT-RBD complex | | Descriptor: | 123-316 scDb, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Jia, G.W, Tong, Z, Tong, J.Y, Su, Z.M. | | Deposit date: | 2024-01-23 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
1A0Q
 
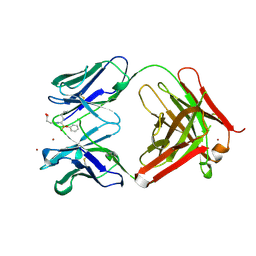 | | 29G11 COMPLEXED WITH PHENYL [1-(1-N-SUCCINYLAMINO)PENTYL] PHOSPHONATE | | Descriptor: | 29G11 FAB (HEAVY CHAIN), 29G11 FAB (LIGHT CHAIN), PHENYL[1-(N-SUCCINYLAMINO)PENTYL]PHOSPHONATE, ... | | Authors: | Buchbinder, J.L, Stephenson, R.C, Scanlan, T.S, Fletterick, R.J. | | Deposit date: | 1997-12-05 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A comparison of the crystallographic structures of two catalytic antibodies with esterase activity.
J.Mol.Biol., 282, 1998
|
|
1AZY
 
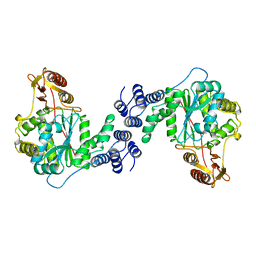 | | STRUCTURAL AND THEORETICAL STUDIES SUGGEST DOMAIN MOVEMENT PRODUCES AN ACTIVE CONFORMATION OF THYMIDINE PHOSPHORYLASE | | Descriptor: | THYMIDINE PHOSPHORYLASE | | Authors: | Pugmire, M.J, Cook, W.J, Jasanoff, A, Walter, M.R, Ealick, S.E. | | Deposit date: | 1997-11-24 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and theoretical studies suggest domain movement produces an active conformation of thymidine phosphorylase.
J.Mol.Biol., 281, 1998
|
|
1D7E
 
 | | CRYSTAL STRUCTURE OF THE P65 CRYSTAL FORM OF PHOTOACTIVE YELLOW PROTEIN | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 1999-10-17 | | Release date: | 2000-03-31 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Conformational substates in different crystal forms of the photoactive yellow protein--correlation with theoretical and experimental flexibility.
Protein Sci., 9, 2000
|
|
7M8W
 
 | | XFEL crystal structure of the prostaglandin D2 receptor CRTH2 in complex with 15R-methyl-PGD2 | | Descriptor: | 15R-methyl-prostaglandin D2, CITRATE ANION, Prostaglandin D2 receptor 2, ... | | Authors: | Shiriaeva, A, Han, G.W, Cherezov, V. | | Deposit date: | 2021-03-30 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Molecular basis for lipid recognition by the prostaglandin D 2 receptor CRTH2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1AZN
 
 | | CRYSTAL STRUCTURE OF THE AZURIN MUTANT PHE114ALA FROM PSEUDOMONAS AERUGINOSA AT 2.6 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Tsai, L.-C, Sjolin, L, Langer, V, Pascher, T, Nar, H. | | Deposit date: | 1994-05-27 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the azurin mutant Phe114Ala from Pseudomonas aeruginosa at 2.6 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1AZU
 
 | |
1AUP
 
 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | NAD-SPECIFIC GLUTAMATE DEHYDROGENASE | | Authors: | Baker, P.J, Waugh, M.L, Stillman, T.J, Turnbull, A.P, Rice, D.W. | | Deposit date: | 1997-09-01 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determinants of substrate specificity in the superfamily of amino acid dehydrogenases.
Biochemistry, 36, 1997
|
|
1E5Z
 
 | |
1E67
 
 | | Zn-Azurin from Pseudomonas aeruginosa | | Descriptor: | AZURIN, NITRATE ION, ZINC ION | | Authors: | Nar, H, Messerschmidt, A. | | Deposit date: | 2000-08-09 | | Release date: | 2000-08-16 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterization and Crystal Structure of Zinc Azurin, a by-Product of Heterologous Expression in Escherichia Coli of Pseudomonas Aeruginosa Copper Azurin
Eur.J.Biochem., 205, 1992
|
|
1E5Y
 
 | |
4HR3
 
 | |
4HWT
 
 | |
4I1Y
 
 | |
