1F58
 
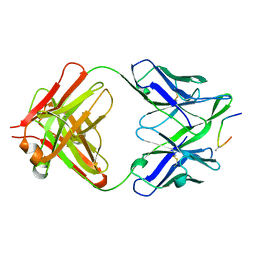 | | IGG1 FAB FRAGMENT (58.2) COMPLEX WITH 24-RESIDUE PEPTIDE (RESIDUES 308-333 OF HIV-1 GP120 (MN ISOLATE) WITH ALA TO AIB SUBSTITUTION AT POSITION 323 | | Descriptor: | Envelope glycoprotein gp120, PROTEIN (IGG1 ANTIBODY 58.2 (HEAVY CHAIN)), PROTEIN (IGG1 ANTIBODY 58.2 (LIGHT CHAIN)) | | Authors: | Stanfield, R.L, Cabezas, E, Satterthwait, A.C, Stura, E.A, Profy, A.T, Wilson, I.A. | | Deposit date: | 1998-10-21 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dual conformations for the HIV-1 gp120 V3 loop in complexes with different neutralizing fabs.
Structure Fold.Des., 7, 1999
|
|
6MB3
 
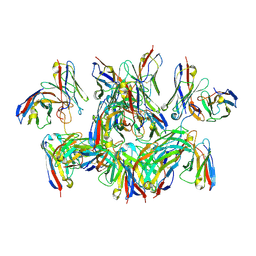 | |
6MHG
 
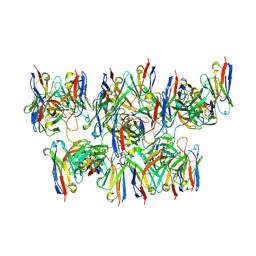 | |
1WCJ
 
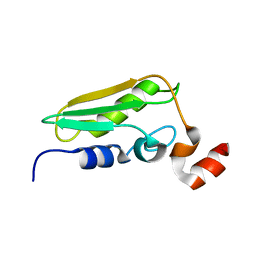 | | Conserved Hypothetical Protein TM0487 from Thermotoga maritima | | Descriptor: | HYPOTHETICAL PROTEIN TM0487 | | Authors: | Almeida, M.S, Peti, W, Herrmann, T, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2004-11-17 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Conserved Hypothetical Protein Tm0487 from Thermotoga Maritima: Implications for 216 Homologous Duf59 Proteins.
Protein Sci., 14, 2005
|
|
6MLM
 
 | | H7 HA0 in complex with Fv from H7.5 IgG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain Fv of H7.5 Fab, Hemagglutinin HA1 chain, ... | | Authors: | Pallesen, J, Turner, H.L, Ward, A.B. | | Deposit date: | 2018-09-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Potent anti-influenza H7 human monoclonal antibody induces separation of hemagglutinin receptor-binding head domains.
PLoS Biol., 17, 2019
|
|
2JZD
 
 | | NMR structure of the domain 527-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
6NF5
 
 | | BG505 MD64 N332-GT5 SOSIP trimer in complex with BG18-like precursor HMP1 fragmentantigen binding and base-binding RM20A3 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG18-like precursor HMP1 fragment antigen binding heavy chain, ... | | Authors: | Ozorowski, G, Torres, J.L, Steichen, J.M, Schief, W.R, Ward, A.B. | | Deposit date: | 2018-12-18 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses.
Science, 366, 2019
|
|
1VK9
 
 | |
6NFC
 
 | | BG505 MD64 N332-GT5 SOSIP trimer in complex with BG18-like precursor HMP42 fragmentantigen binding and base-binding RM20A3 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG18-like precursor HMP42 fragment antigen binding heavy chain, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2018-12-19 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses.
Science, 366, 2019
|
|
6NC3
 
 | | AMC011 v4.2 SOSIP Env trimer in complex with fusion peptide targeting antibody VRC34 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Env AMC011 v4.2 SOSIP gp120, ... | | Authors: | Cottrell, C.A, Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2018-12-10 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Conformational Plasticity in the HIV-1 Fusion Peptide Facilitates Recognition by Broadly Neutralizing Antibodies.
Cell Host Microbe, 25, 2019
|
|
1VK8
 
 | |
1VLR
 
 | |
1VQ3
 
 | |
1VKH
 
 | |
1VJO
 
 | |
1VR3
 
 | |
1VRM
 
 | |
1VKM
 
 | |
1VQ0
 
 | |
1VJ1
 
 | |
2JZE
 
 | | NMR structure of the domain 527-651 of the SARS-CoV nonstructural protein nsp3, single conformer closest to the mean coordinates of an ensemble of twenty energy minimized conformers | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
1VJ2
 
 | |
1VKB
 
 | |
1VLQ
 
 | |
1VKY
 
 | |
