7X70
 
 | |
7X6Y
 
 | |
6BTI
 
 | | Crystal structure of human cellular retinol binding protein 2 (CRBP2) in complex with N-arachidonoylethanolamine (AEA) | | Descriptor: | (5Z,8Z,11Z,14Z)-N-(2-hydroxyethyl)icosa-5,8,11,14-tetraenamide, DI(HYDROXYETHYL)ETHER, Retinol-binding protein 2 | | Authors: | Silvaroli, J.A, Blaner, W.S, Lodowski, D.T, Golczak, M. | | Deposit date: | 2017-12-06 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Retinol-binding protein 2 (RBP2) binds monoacylglycerols and modulates gut endocrine signaling and body weight.
Sci Adv, 6, 2020
|
|
6BTH
 
 | | Crystal structure of human cellular retinol binding protein 2 (CRBP2) in complex with 2-arachidonoylglycerol (2-AG) | | Descriptor: | 1,3-dihydroxypropan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, DI(HYDROXYETHYL)ETHER, Retinol-binding protein 2 | | Authors: | Silvaroli, J.A, Blaner, W.S, Lodowski, D.T, Golczak, M. | | Deposit date: | 2017-12-06 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Retinol-binding protein 2 (RBP2) binds monoacylglycerols and modulates gut endocrine signaling and body weight.
Sci Adv, 6, 2020
|
|
6BUG
 
 | |
2O3H
 
 | | Crystal structure of the human C65A Ape | | Descriptor: | ACETATE ION, DNA-(apurinic or apyrimidinic site) lyase, SAMARIUM (III) ION | | Authors: | Georgiadis, M.M, Gaur, R.K, Delaplane, S, Svenson, J. | | Deposit date: | 2006-12-01 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease
Mutat.Res., 643, 2008
|
|
6IMJ
 
 | | The crystal structure of Se-AsfvLIG:DNA complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Chen, Y.Q, Gan, J.H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-02-27 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Structure of the error-prone DNA ligase of African swine fever virus identifies critical active site residues.
Nat Commun, 10, 2019
|
|
2NWM
 
 | |
2O3C
 
 | | Crystal structure of zebrafish Ape | | Descriptor: | APEX nuclease 1, LEAD (II) ION | | Authors: | Georgiadis, M.M, Gaur, R.K, Delaplane, S, Svenson, J. | | Deposit date: | 2006-12-01 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease
Mutat.Res., 643, 2008
|
|
6DWW
 
 | | Hermes transposase deletion dimer complex with (A/T) DNA and Mn2+ | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*GP*AP*GP*AP*AP*CP*AP*AP*CP*AP*AP*CP*AP*AP*G)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*A)-3'), ... | | Authors: | Dyda, F, Voth, A.R, Hickman, A.B. | | Deposit date: | 2018-06-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Structural insights into the mechanism of double strand break formation by Hermes, a hAT family eukaryotic DNA transposase.
Nucleic Acids Res., 46, 2018
|
|
5UXZ
 
 | |
5WTK
 
 | | Crystal structure of RNP complex | | Descriptor: | CRISPR-associated endoribonuclease C2c2, RNA (58-MER) | | Authors: | Liu, L, Wang, Y. | | Deposit date: | 2016-12-13 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Two Distant Catalytic Sites Are Responsible for C2c2 RNase Activities
Cell, 168, 2017
|
|
5UY1
 
 | | X-ray crystal structure of apo Halotag | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Dunham, N.P, Boal, A.K. | | Deposit date: | 2017-02-23 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Cation-pi Interaction Enables a Halo-Tag Fluorogenic Probe for Fast No-Wash Live Cell Imaging and Gel-Free Protein Quantification.
Biochemistry, 56, 2017
|
|
5WTJ
 
 | | Crystal structure of an endonuclease | | Descriptor: | CRISPR-associated endoribonuclease C2c2 | | Authors: | Liu, L, Wang, Y. | | Deposit date: | 2016-12-13 | | Release date: | 2017-02-08 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Two Distant Catalytic Sites Are Responsible for C2c2 RNase Activities
Cell, 168, 2017
|
|
2M8T
 
 | |
8HLP
 
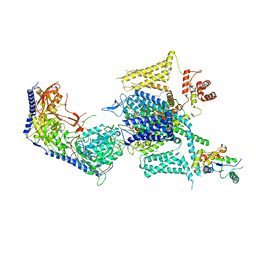 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 (apo) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wei, Y, Yu, Z, Zhao, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
7EPX
 
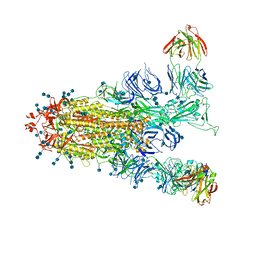 | | S protein of SARS-CoV-2 in complex with GW01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Shen, Y.P, Zhang, Y.Y, Yan, R.H, Li, Y.N, Zhou, Q. | | Deposit date: | 2021-04-28 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Novel sarbecovirus bispecific neutralizing antibodies with exceptional breadth and potency against currently circulating SARS-CoV-2 variants and sarbecoviruses.
Cell Discov, 8, 2022
|
|
8HMB
 
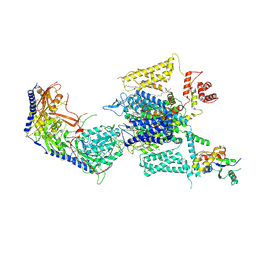 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 in complex with benidipine (BEN) | | Descriptor: | (3R)-1-benzylpiperidin-3-yl methyl (2R,3R,4R,5R,6S)-2,6-dimethyl-4-(3-nitrophenyl)piperidine-3,5-dicarboxylate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wei, Y, Yu, Z, Zhao, Y. | | Deposit date: | 2022-12-02 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
8HMA
 
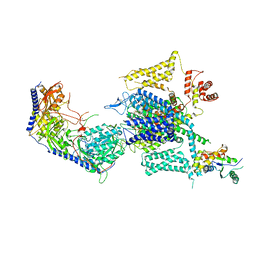 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 in complex with tetrandrine (TET) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6,6',7,12-tetramethoxy-2,2'-dimethyl-1beta-3,4-didehydroberbaman, ... | | Authors: | Wei, Y, Yu, Z, Zhao, Y. | | Deposit date: | 2022-12-02 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
6M1H
 
 | | CryoEM structure of human PAC1 receptor in complex with maxadilan | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
6M1I
 
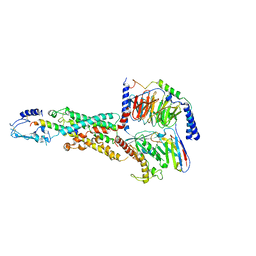 | | CryoEM structure of human PAC1 receptor in complex with PACAP38 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
3ONL
 
 | | yeast Ent3_ENTH-Vti1p_Habc complex structure | | Descriptor: | Epsin-3, t-SNARE VTI1 | | Authors: | Wang, J, Fang, P, Niu, L, Teng, M. | | Deposit date: | 2010-08-29 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Epsin N-terminal homology domains bind on opposite sides of two SNAREs
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3ONK
 
 | | yeast Ent3_ENTH domain | | Descriptor: | Epsin-3 | | Authors: | Wang, J, Fang, P, Niu, L, Teng, M. | | Deposit date: | 2010-08-29 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Epsin N-terminal homology domains bind on opposite sides of two SNAREs
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3ONJ
 
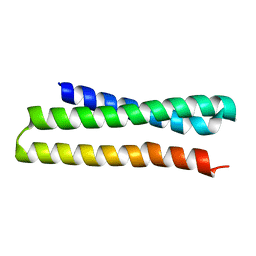 | |
8J0D
 
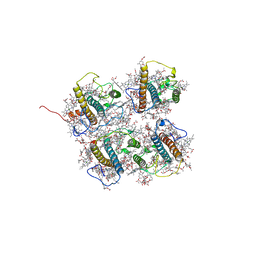 | | FCP heterodimer, Lhca2, and Lhcf5 together as the M1 side binds to the PSII core in the diatom Thalassiosira pseudonana | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Li, Z, Feng, Y, Wang, W, Shen, J.R. | | Deposit date: | 2023-04-10 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structure of a diatom photosystem II supercomplex containing a member of Lhcx family and dimeric FCPII.
Sci Adv, 9, 2023
|
|
