3VTB
 
 | | Crystal structure of rat vitamin D receptor bound to a partial agonist 25-adamantyl-23-yne-19-norvitammin D ADTK1 | | Descriptor: | (1R,3R,7E,17beta)-17-{(2R,6S)-6-hydroxy-6-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]hex-4-yn-2-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, COACTIVATOR PEPTIDE DRIP, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Kudo, T, Tokiwa, H, Makishima, M, Yamada, S, Ikura, T, Ito, N. | | Deposit date: | 2012-05-26 | | Release date: | 2013-06-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Combination of Triple Bond and Adamantane Ring on the Vitamin D Side Chain Produced Partial Agonists for Vitamin D Receptor.
J.Med.Chem., 2014
|
|
3IFB
 
 | | NMR STUDY OF HUMAN INTESTINAL FATTY ACID BINDING PROTEIN | | Descriptor: | INTESTINAL FATTY ACID BINDING PROTEIN | | Authors: | Zhang, F, Luecke, C, Baier, L.J, Sacchettini, J.C, Hamilton, J.A. | | Deposit date: | 1998-10-16 | | Release date: | 1998-10-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human intestinal fatty acid binding protein: implications for ligand entry and exit.
J.Biomol.NMR, 9, 1997
|
|
3VTC
 
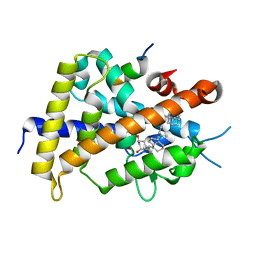 | | Crystal structure of rat vitamin D receptor bound to a partial agonist 26-adamantyl-23-yne-19-norvitammin D ADTK3 | | Descriptor: | (1R,3R,7E,17beta)-17-{(2R,6R)-6-hydroxy-7-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]hept-4-yn-2-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, 1,2-ETHANEDIOL, COACTIVATOR PEPTIDE DRIP, ... | | Authors: | Nakabayashi, M, Kudo, T, Tokiwa, H, Makishima, M, Yamada, S, Ikura, T, Ito, N. | | Deposit date: | 2012-05-26 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Combination of Triple Bond and Adamantane Ring on the Vitamin D Side Chain Produced Partial Agonists for Vitamin D Receptor.
J.Med.Chem., 2014
|
|
5DGR
 
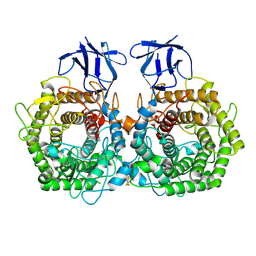 | | Crystal structure of GH9 exo-beta-D-glucosaminidase PBPRA0520, glucosamine complex | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, Putative endoglucanase-related protein, SODIUM ION | | Authors: | Suzuki, K, Honda, Y, Fushinobu, S. | | Deposit date: | 2015-08-28 | | Release date: | 2015-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of an inverting glycoside hydrolase family 9 exo-beta-D-glucosaminidase and the design of glycosynthase.
Biochem.J., 473, 2016
|
|
6I8L
 
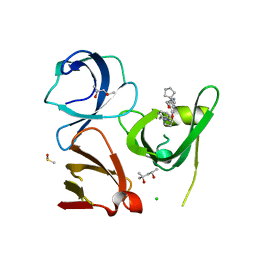 | | Crystal structure of Spindlin1 in complex with the inhibitor TD001851a | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 5'-(cyclopropylmethoxy)-6'-[3-(1,3-dihydroisoindol-2-yl)propoxy]spiro[cyclopentane-1,3'-indole]-2'-amine, ... | | Authors: | Johansson, C, Fagan, V, Brennan, P.E, Sorrell, F.J, Krojer, T, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
5DGQ
 
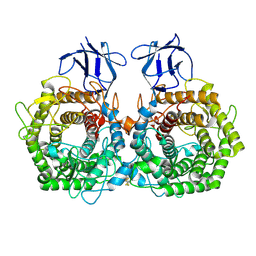 | |
3VTD
 
 | | Crystal structure of rat vitamin D receptor bound to a partial agonist 26-adamantyl-23-yne-19-norvitammin D ADTK4 | | Descriptor: | (1R,3R,7E,17beta)-17-{(2R,6S)-6-hydroxy-7-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]hept-4-yn-2-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, COACTIVATOR PEPTIDE DRIP, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Kudo, T, Tokiwa, H, Makishima, M, Yamada, S, Ikura, T, Ito, N. | | Deposit date: | 2012-05-26 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Combination of Triple Bond and Adamantane Ring on the Vitamin D Side Chain Produced Partial Agonists for Vitamin D Receptor.
J.Med.Chem., 2014
|
|
6JWH
 
 | | Yeast Npl4 zinc finger, MPN and CTD domains | | Descriptor: | GLYCEROL, Nuclear protein localization protein 4, ZINC ION | | Authors: | Sato, Y, Fukai, S. | | Deposit date: | 2019-04-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.72000253 Å) | | Cite: | Structural insights into ubiquitin recognition and Ufd1 interaction of Npl4.
Nat Commun, 10, 2019
|
|
3W32
 
 | | EGFR kinase domain complexed with compound 20a | | Descriptor: | 4-({3-chloro-4-[3-(trifluoromethyl)phenoxy]phenyl}amino)-N-[2-(methylsulfonyl)ethyl]-8,9-dihydro-7H-pyrimido[4,5-b]azepine-6-carboxamide, Epidermal growth factor receptor, SULFATE ION | | Authors: | Sogabe, S, Kawakita, Y. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and synthesis of novel pyrimido[4,5-b]azepine derivatives as HER2/EGFR dual inhibitors
Bioorg.Med.Chem., 21, 2013
|
|
6HUS
 
 | | 2'-fucosyllactose and 3-fucosyllactose binding protein from Bifidobacterium longum infantis, bound with 3-fucosyllactose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ABC transporter substrate-binding protein, ZINC ION, ... | | Authors: | Ejby, M, Abou Hachem, M, Lo Leggio, L, Takane, K, Sakanaka, M. | | Deposit date: | 2018-10-09 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.409 Å) | | Cite: | Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis.
Sci Adv, 5, 2019
|
|
6I8B
 
 | | Crystal structure of Spindlin1 in complex with the inhibitor VinSpinIn | | Descriptor: | 2-[4-[2-[[2-[3-[2-azanyl-5-(cyclopropylmethoxy)-3,3-dimethyl-indol-6-yl]oxypropyl]-1,3-dihydroisoindol-5-yl]oxy]ethyl]-1,2,3-triazol-1-yl]-1-[4-(2-pyrrolidin-1-ylethyl)piperidin-1-yl]ethanone, DIMETHYL SULFOXIDE, GLYCINE, ... | | Authors: | Johansson, C, Fagan, V, Brennan, P.E, Sorrell, F.J, Krojer, T, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
6I8Y
 
 | | Crystal structure of Spindlin1 in complex with the Methyltransferase inhibitor A366 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 5'-methoxy-6'-[3-(pyrrolidin-1-yl)propoxy]spiro[cyclobutane-1,3'-indol]-2'-amine, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Shrestha, L, Sorrell, F.J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-21 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
3VMH
 
 | | Oxygen-bound complex between oxygenase and ferredoxin in carbazole 1,9a-dioxygenase | | Descriptor: | FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin component of carbazole, ... | | Authors: | Ashikawa, Y, Nojiri, H. | | Deposit date: | 2011-12-12 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insight into the substrate- and dioxygenbinding manner in the catalytic cycle of rieske nonheme iron oxygenase system, carbazole 1,9adioxygenase
Bmc Struct.Biol., 12, 2012
|
|
3VMG
 
 | | Reduced carbazole-bound complex between oxygenase and ferredoxin in carbazole 1,9a-dioxygenase | | Descriptor: | 9H-CARBAZOLE, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ashikawa, Y, Nojiri, H. | | Deposit date: | 2011-12-12 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insight into the substrate- and dioxygenbinding manner in the catalytic cycle of rieske nonheme iron oxygenase system, carbazole 1,9adioxygenase
Bmc Struct.Biol., 12, 2012
|
|
6HUR
 
 | | 2'-fucosyllactose and 3-fucosyllactose binding protein from Bifidobacterium longum infantis, bound with 2'-fucosyllactose | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ABC transporter substrate-binding protein, ... | | Authors: | Ejby, M, Abou Hachem, M, Lo Leggio, L, Katayama, T, Sakanaka, M. | | Deposit date: | 2018-10-09 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.297 Å) | | Cite: | Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis.
Sci Adv, 5, 2019
|
|
2ZYE
 
 | | Structure of HIV-1 Protease in Complex with Potent Inhibitor KNI-272 Determined by Neutron Crystallography | | Descriptor: | (4R)-N-tert-butyl-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl-L-cysteinyl}amino)-4-phenylbutanoyl]-1,3-thiazolidine-4-carboxamide, protease | | Authors: | Adachi, M, Ohhara, T, Tamada, T, Okazaki, N, Kuroki, R. | | Deposit date: | 2009-01-20 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-29 | | Method: | NEUTRON DIFFRACTION (1.9 Å) | | Cite: | Structure of HIV-1 protease in complex with potent inhibitor KNI-272 determined by high-resolution X-ray and neutron crystallography.
Proc.Natl.Acad.Sci.USA, 2009
|
|
6JWJ
 
 | | Npl4 in complex with Ufd1 | | Descriptor: | GLYCEROL, Nuclear protein localization protein 4, Peptide from Ubiquitin fusion degradation protein 1, ... | | Authors: | Sato, Y, Fukai, S. | | Deposit date: | 2019-04-20 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural insights into ubiquitin recognition and Ufd1 interaction of Npl4.
Nat Commun, 10, 2019
|
|
3W5M
 
 | | Crystal Structure of Streptomyces avermitilis alpha-L-rhamnosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Putative rhamnosidase | | Authors: | Fujimoto, Z, Jackson, A, Michikawa, M, Maehara, T, Momma, M, Henrissat, B.F, Gilbert, H.J, Kaneko, S. | | Deposit date: | 2013-01-31 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a Streptomyces avermitilis alpha-L-rhamnosidase reveals a novel carbohydrate-binding module CBM67 within the six-domain arrangement.
J.Biol.Chem., 288, 2013
|
|
3ATK
 
 | | Crystal structure of trypsin complexed with cycloheptanamine | | Descriptor: | CALCIUM ION, Cationic trypsin, DIMETHYL SULFOXIDE, ... | | Authors: | Yamane, J, Yao, M, Tanaka, I. | | Deposit date: | 2011-01-05 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3ATI
 
 | | Crystal structure of trypsin complexed with (3-methoxyphenyl)methanamine | | Descriptor: | 1-(3-methoxyphenyl)methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Tanaka, I. | | Deposit date: | 2011-01-05 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
2E7S
 
 | |
3AQO
 
 | |
3ATM
 
 | | Crystal structure of trypsin complexed with 2-(1H-indol-3-yl)ethanamine | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANAMINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Tanaka, I. | | Deposit date: | 2011-01-05 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3ATL
 
 | | Crystal structure of trypsin complexed with benzamidine | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Tanaka, I. | | Deposit date: | 2011-01-05 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3W5N
 
 | | Crystal Structure of Streptomyces avermitilis alpha-L-rhamnosidase complexed with L-rhamnose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Putative rhamnosidase, ... | | Authors: | Fujimoto, Z, Jackson, A, Michikawa, M, Maehara, T, Momma, M, Henrissat, B.F, Gilbert, H.J, Kaneko, S. | | Deposit date: | 2013-01-31 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a Streptomyces avermitilis alpha-L-rhamnosidase reveals a novel carbohydrate-binding module CBM67 within the six-domain arrangement.
J.Biol.Chem., 288, 2013
|
|
