2MTF
 
 | | Solution structure of the La motif of human LARP6 | | Descriptor: | La-related protein 6 | | Authors: | Martino, L, Salisbury, N.JH, Atkinson, A.R, Kelly, G, Conte, M.R. | | Deposit date: | 2014-08-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module.
Nucleic Acids Res., 43, 2015
|
|
5EB3
 
 | | VB6-bound protein | | Descriptor: | 4,5-bis(hydroxymethyl)-2-methyl-pyridin-3-ol, SULFATE ION, YfiR | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
5EBB
 
 | | Structure of human sphingomyelinase phosphodiesterase like 3A (SMPDL3A) with Zn2+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, GLYCEROL, ... | | Authors: | Lim, S.M, Yeung, K, Tresaugues, L, Teo, H.L, Nordlund, P. | | Deposit date: | 2015-10-19 | | Release date: | 2016-01-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure and catalytic mechanism of human sphingomyelin phosphodiesterase like 3a - an acid sphingomyelinase homologue with a novel nucleotide hydrolase activity.
Febs J., 283, 2016
|
|
8Q1Q
 
 | | mouse Keap1 in complex with stapled peptide | | Descriptor: | Kelch-like ECH-associated protein 1, SULFATE ION, Stapled peptide, ... | | Authors: | Kack, H, Wissler, L. | | Deposit date: | 2023-08-01 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.378 Å) | | Cite: | A cell-active cyclic peptide targeting the Nrf2/Keap1 protein-protein interaction.
Chem Sci, 14, 2023
|
|
6D02
 
 | | Cross-alpha Amyloid-like Structure alphaAmL, 2nd form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CARBONATE ION, PHOSPHATE ION, ... | | Authors: | Zhang, S.-Q, Liu, L, Degrado, W.F. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
5DWV
 
 | | Crystal structure of the Luciferase complexed with substrate analogue | | Descriptor: | 2-[6-(cyclobuta-1,3-dien-1-ylamino)-1,3-benzothiazol-2-yl]-1,3-thiazol-4-ol, Luciferin 4-monooxygenase | | Authors: | Su, J, Li, Z, Yuan, Z, Gu, L. | | Deposit date: | 2015-09-23 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Luciferase complexed with substrate analogue
To Be Published
|
|
5DP6
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 7 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[N-(3-cyclopropylpropanoyl)-L-phenylalanyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
6CR2
 
 | | Crystal structure of sterol 14-alpha demethylase (CYP51B) from Aspergillus fumigatus in complex with the VNI derivative N-(1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl)-4-(5-(2-fluoro-4-(2,2,2-trifluoroethoxy)phenyl)-1,3,4-oxadiazol-2-yl)benzamide | | Descriptor: | 14-alpha sterol demethylase Cyp51B, N-[(1R)-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl]-4-{5-[2-fluoro-4-(2,2,2-trifluoroethoxy)phenyl]-1,3,4-oxadiazol-2-yl}benzamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Friggeri, L, Hargrove, T.Y, Wawrzak, Z, Lepesheva, G.I. | | Deposit date: | 2018-03-16 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Sterol 14 alpha-Demethylase Structure-Based Design of VNI (( R)- N-(1-(2,4-Dichlorophenyl)-2-(1 H-imidazol-1-yl)ethyl)-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide)) Derivatives To Target Fungal Infections: Synthesis, Biological Evaluation, and Crystallographic Analysis.
J. Med. Chem., 61, 2018
|
|
8Q1R
 
 | | mouse Keap1 in complex with stapled peptide | | Descriptor: | Kelch-like ECH-associated protein 1, SODIUM ION, SULFATE ION, ... | | Authors: | Kack, H, Wissler, L. | | Deposit date: | 2023-08-01 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | A cell-active cyclic peptide targeting the Nrf2/Keap1 protein-protein interaction.
Chem Sci, 14, 2023
|
|
5DY2
 
 | | Crystal structure of Dbr2 with mutation M27L | | Descriptor: | 1,6-DIHYDROXY NAPHTHALENE, Artemisinic aldehyde Delta(11(13)) reductase, FLAVIN MONONUCLEOTIDE | | Authors: | Li, L, Chen, T.T, Xu, Y.C. | | Deposit date: | 2015-09-24 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The Crystal structure of Dbr2
to be published
|
|
1RFY
 
 | | Crystal Structure of Quorum-Sensing Antiactivator TraM | | Descriptor: | Transcriptional repressor traM | | Authors: | Chen, G, Malenkos, J.W, Cha, M.R, Fuqua, C, Chen, L. | | Deposit date: | 2003-11-10 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Quorum-sensing antiactivator TraM forms a dimer that dissociates to inhibit TraR
Mol.Microbiol., 52, 2004
|
|
8PBY
 
 | |
8PC1
 
 | |
1VJK
 
 | | Putative molybdopterin converting factor, subunit 1 from Pyrococcus furiosus, Pfu-562899-001 | | Descriptor: | molybdopterin converting factor, subunit 1 | | Authors: | Chen, L, Liu, Z.J, Tempel, W, Shah, A, Lee, D, Rose, J.P, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Putative molybdopterin converting factor, subunit 1 from Pyrococcus furiosus, Pfu-562899-001 '
To be published
|
|
1S1J
 
 | | Crystal Structure of ZipA in complex with indoloquinolizin inhibitor 1 | | Descriptor: | (12bS)-1,2,3,4,12,12b-hexahydroindolo[2,3-a]quinolizin-7(6H)-one, Cell division protein zipA | | Authors: | Jenning, L.D, Foreman, K.W, Rush III, T.S, Tsao, D.H, Mosyak, L, Li, Y, Sukhdeo, M.N, Ding, W, Dushin, E.G, Kenney, C.H, Moghazeh, S.L, Peterson, P.J, Ruzin, A.V, Tuckman, M, Sutherland, A.G. | | Deposit date: | 2004-01-06 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Design and synthesis of indolo[2,3-a]quinolizin-7-one inhibitors of the ZipA-FtsZ interaction
Bioorg.Med.Chem.Lett., 14, 2004
|
|
8PBZ
 
 | |
8PBX
 
 | |
8PC0
 
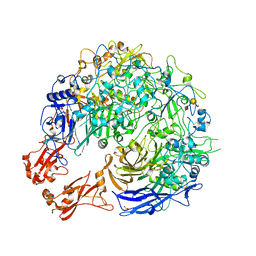 | | Sub-tomogram average of the open conformation of the Nap adhesion complex from the human pathogen Mycoplasma genitalium. | | Descriptor: | Adhesin P1, Mgp-operon protein 3, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-1,5-anhydro-D-glucitol, ... | | Authors: | Sprankel, L, Scheffer, M.P, Frangakis, A.F. | | Deposit date: | 2023-06-09 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Cryo-electron tomography reveals the binding and release states of the major adhesion complex from Mycoplasma genitalium.
Plos Pathog., 19, 2023
|
|
1S7H
 
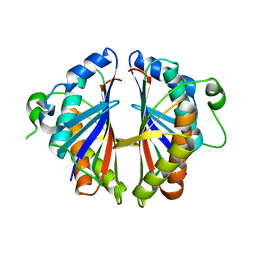 | | Structural Genomics, 2.2A crystal structure of protein YKOF from Bacillus subtilis | | Descriptor: | ykoF | | Authors: | Zhang, R, Lezondra, L, Moy, S, Dementieva, I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-01-29 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.2A crystal structure of protein YKOF from Bacillus subtilis
To be Published
|
|
6CTY
 
 | | Crystal structure of dihydroorotase pyrC from Yersinia pestis in complex with zinc and malate at 2.4 A resolution | | Descriptor: | D-MALATE, Dihydroorotase, ZINC ION | | Authors: | Lipowska, J, Shabalin, I.G, Winsor, J, Woinska, M, Cooper, D.R, Kwon, K, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-03-23 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Pyrimidine biosynthesis in pathogens - Structures and analysis of dihydroorotases from Yersinia pestis and Vibrio cholerae.
Int.J.Biol.Macromol., 136, 2019
|
|
5DU2
 
 | | Structural analysis of EspG2 glycosyltransferase | | Descriptor: | EspG2 glycosyltransferase | | Authors: | Michalska, K, Elshahawi, S.I, Bigelow, L, Babnigg, G, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-09-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of EspG2 glycosyltransferase
To Be Published
|
|
8Q2E
 
 | | The 1.68-A X-ray crystal structure of Sporosarcina pasteurii urease inhibited by thiram and bound to dimethylditiocarbamate | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Ciurli, S. | | Deposit date: | 2023-08-02 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Kinetic and structural details of urease inactivation by thiuram disulphides.
J.Inorg.Biochem., 250, 2023
|
|
6CYN
 
 | | CTX-M-14 N106S/D240G mutant | | Descriptor: | Beta-lactamase | | Authors: | Patel, M.P, Hu, L, Sankaran, B, Brown, C, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2018-04-06 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Synergistic effects of functionally distinct substitutions in beta-lactamase variants shed light on the evolution of bacterial drug resistance.
J. Biol. Chem., 293, 2018
|
|
1S8C
 
 | | Crystal structure of human heme oxygenase in a complex with biliverdine | | Descriptor: | BILIVERDINE IX ALPHA, Heme oxygenase 1 | | Authors: | Lad, L, Friedman, J, Li, H, Bhaskar, B, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2004-02-02 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of Human Heme Oxygenase-1 in a Complex with Biliverdin
Biochemistry, 43, 2004
|
|
1S9D
 
 | | ARF1[DELTA 1-17]-GDP-MG IN COMPLEX WITH BREFELDIN A AND A SEC7 DOMAIN | | Descriptor: | 1,6,7,8,9,11A,12,13,14,14A-DECAHYDRO-1,13-DIHYDROXY-6-METHYL-4H-CYCLOPENT[F]OXACYCLOTRIDECIN-4-ONE, ADP-Ribosylation Factor 1, Arno, ... | | Authors: | Renault, L, Guibert, B, Cherfils, J. | | Deposit date: | 2004-02-04 | | Release date: | 2004-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Snapshots of the Mechanism and Inhibition of a Guanine Nucleotide Exchange Factor
Nature, 426, 2003
|
|
