3RMS
 
 | | Crystal structure of uncharacterized protein Svir_20580 from Saccharomonospora viridis | | Descriptor: | GLYCEROL, ZINC ION, uncharacterized protein | | Authors: | Michalska, K, Weger, A, Hatzos-Skintges, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-21 | | Release date: | 2011-05-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Crystal structure of uncharacterized protein Svir_20580 from Saccharomonospora viridis
To be Published
|
|
4MV2
 
 | | Crystal structure of plu4264 protein from Photorhabdus luminescens | | Descriptor: | NICKEL (II) ION, SODIUM ION, plu4264 | | Authors: | Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Thomas, M.G, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
4MY0
 
 | | Crystal Structure of GCN5-related N-acetyltransferase from Kribbella flavida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYL COENZYME *A, GCN5-related N-acetyltransferase, ... | | Authors: | Kim, Y, Mack, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-26 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Crystal Structure of GCN5-related N-acetyltransferase from Kribbella flavida
To be Published
|
|
3HQX
 
 | | Crystal structure of protein of unknown function (DUF1255,PF06865) from Acinetobacter sp. ADP1 | | Descriptor: | UPF0345 protein ACIAD0356 | | Authors: | Nocek, B, Hatzos, C, Freeman, L, Dauter, Z, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-06-08 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of protein of unknown function (DUF1255,PF06865) from Acinetobacter sp. ADP1
To be Published
|
|
3I4P
 
 | | Crystal structure of AsnC family transcriptional regulator from Agrobacterium tumefaciens | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Cymborowski, M, Chruszcz, M, Luo, H, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-02 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of AsnC family transcriptional regulator from Agrobacterium tumefaciens
To be Published
|
|
3RAO
 
 | | Crystal Structure of the Luciferase-like Monooxygenase from Bacillus cereus ATCC 10987. | | Descriptor: | Putative Luciferase-like Monooxygenase, SULFATE ION | | Authors: | Domagalski, M.J, Chruszcz, M, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-28 | | Release date: | 2011-05-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Luciferase-like Monooxygenase from Bacillus cereus ATCC 10987.
To be Published
|
|
3IUV
 
 | | The structure of a member of TetR family (SCO1917) from Streptomyces coelicolor A3 | | Descriptor: | uncharacterized TetR family protein | | Authors: | Tan, K, Cuff, M, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | The structure of a member of TetR family (SCO1917) from Streptomyces coelicolor A3
To be Published
|
|
1KUT
 
 | | Structural Genomics, Protein TM1243, (SAICAR synthetase) | | Descriptor: | Phosphoribosylaminoimidazole-succinocarboxamide synthase | | Authors: | Zhang, R, Skarina, T, Beasley, S, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-22 | | Release date: | 2002-08-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of SAICAR synthase from Thermotoga maritima at 2.2 angstroms reveals an unusual covalent dimer.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1Y89
 
 | | Crystal Structure of devB protein | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, DI(HYDROXYETHYL)ETHER, NONAETHYLENE GLYCOL, ... | | Authors: | Lazarski, K, Cymborowski, M, Chruszcz, M, Zheng, H, Zhang, R, Lezondra, L, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-10 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of devB protein
To be Published
|
|
4PAG
 
 | | ABC transporter solute binding protein from Sulfurospirillum deleyianum DSM 6946 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, HISTIDINE, ... | | Authors: | Chang, C, Endres, M, Li, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-08 | | Release date: | 2014-04-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Crystal structure of ABC transporter solute binding protein from Sulfurospirillum deleyianum DSM 6946
To Be Published
|
|
3K6H
 
 | | Crystal structure of a nitroreductase family protein from Agrobacterium tumefaciens str. C58 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Nitroreductase family protein, SULFATE ION | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-08 | | Release date: | 2009-10-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of a nitroreductase family protein from Agrobacterium tumefaciens str. C58
To be Published
|
|
3U4G
 
 | | The Structure of CobT from Pyrococcus horikoshii | | Descriptor: | ACETIC ACID, NaMN:DMB phosphoribosyltransferase, SULFATE ION | | Authors: | Cuff, M.E, Evdokimova, E, Mursleen, A, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-10-07 | | Release date: | 2011-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of CobT from Pyrococcus horikoshii
TO BE PUBLISHED
|
|
4OVX
 
 | | Crystal structure of Xylose isomerase domain protein from Planctomyces limnophilus DSM 3776 | | Descriptor: | 1,2-ETHANEDIOL, Xylose isomerase domain protein TIM barrel | | Authors: | Chang, C, Bigelow, L, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-22 | | Release date: | 2014-02-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Crystal structure of Xylose isomerase domain protein from Planctomyces limnophilus DSM 3776
To be published
|
|
3GYK
 
 | | The crystal structure of a thioredoxin-like oxidoreductase from Silicibacter pomeroyi DSS-3 | | Descriptor: | 1,2-ETHANEDIOL, 27kDa outer membrane protein, SULFATE ION | | Authors: | Fan, Y, Marshall, N, Keigher, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-03 | | Release date: | 2009-06-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The crystal structure of a thioredoxin-like oxidoreductase from Silicibacter pomeroyi DSS-3
To be Published
|
|
4OG2
 
 | | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with leucine | | Descriptor: | Amino acid/amide ABC transporter substrate-binding protein, HAAT family, CHLORIDE ION, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-15 | | Release date: | 2014-01-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.099 Å) | | Cite: | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with leucine
To be Published
|
|
1XWM
 
 | |
3JU1
 
 | | Crystal Structure of Enoyl-CoA Hydratase/Isomerase Family Protein | | Descriptor: | ACETIC ACID, Enoyl-CoA hydratase/isomerase family protein, FORMIC ACID, ... | | Authors: | Kim, Y, Xu, X, Cui, H, Ng, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal Structure of Enoyl-CoA Hydratase/Isomerase Family Protein
To be Published
|
|
4OAD
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with chloramphenicol | | Descriptor: | 1,2-ETHANEDIOL, CHLORAMPHENICOL, GNAT superfamily acetyltransferase PA4794, ... | | Authors: | Majorek, K.A, Niedzialkowska, E, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-04 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with chloramphenicol
To be Published
|
|
4OBB
 
 | | The crystal structure of a solute-binding protein from Anabaena variabilis ATCC 29413 in complex with (3S)-3-methyl-2-oxopentanoic acid. | | Descriptor: | (3S)-3-methyl-2-oxopentanoic acid, Amino acid/amide ABC transporter substrate-binding protein, HAAT family, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-07 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.526 Å) | | Cite: | The crystal structure of a solute-binding protein from Anabaena variabilis ATCC 29413 in complex with (3S)-3-methyl-2-oxopentanoic acid.
To be Published
|
|
3K6C
 
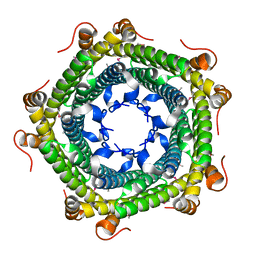 | | Crystal structure of protein ne0167 from nitrosomonas europaea | | Descriptor: | Uncharacterized protein NE0167 | | Authors: | Chang, C, Evdokimova, E, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-08 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Protein Ne0167 from Nitrosomonas Europaea
To be Published
|
|
4OTZ
 
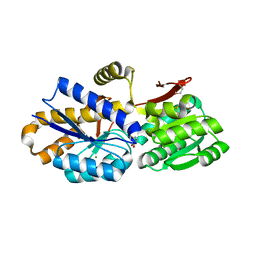 | | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with cystein | | Descriptor: | Amino acid/amide ABC transporter substrate-binding protein, HAAT family, CYSTEINE, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-02-14 | | Release date: | 2014-03-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with cystein
To be Published
|
|
3QOE
 
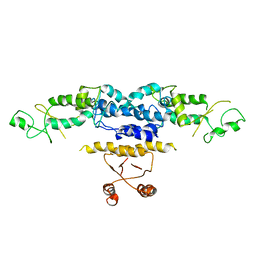 | | Crystal Structure of Heterocyst Differentiation Protein, HetR from Fischerella mv11 | | Descriptor: | Heterocyst differentiation protein | | Authors: | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Structure of transcription factor HetR required for heterocyst differentiation in cyanobacteria.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1PVM
 
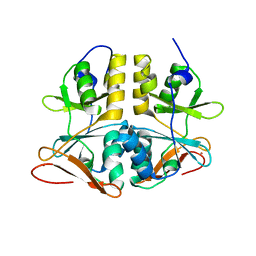 | | Crystal Structure of a Conserved CBS Domain Protein TA0289 of Unknown Function from Thermoplasma acidophilum | | Descriptor: | MERCURY (II) ION, conserved hypothetical protein Ta0289 | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Savchenko, A, Xu, L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-06-27 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biochemical and structural characterization of a novel family of cystathionine beta-synthase domain proteins fused to a Zn ribbon-like domain
J.Mol.Biol., 375, 2008
|
|
3K0B
 
 | | Crystal structure of a predicted N6-adenine-specific DNA methylase from Listeria monocytogenes str. 4b F2365 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, predicted N6-adenine-specific DNA methylase | | Authors: | Nocek, B, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-24 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a predicted N6-adenine-specific DNA methylase from Listeria monocytogenes str. 4b F2365
To be Published
|
|
4QN8
 
 | | The crystal structure of an effector protein VipE from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | BETA-MERCAPTOETHANOL, VipE | | Authors: | Tan, K, Xu, X, Cui, H, Liu, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-16 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | The crystal structure of an effector protein VipE from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To be Published
|
|
