4DDF
 
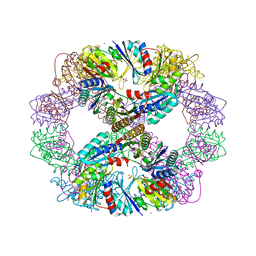 | | Computationally Designed Self-assembling Octahedral Cage protein, O333, Crystallized in space group P4 | | Descriptor: | CHLORIDE ION, Propanediol utilization polyhedral body protein PduT, SULFATE ION | | Authors: | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2012-01-18 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
4EGG
 
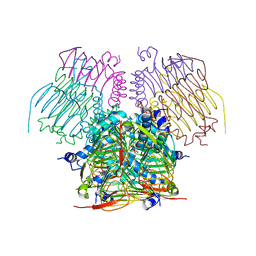 | | Computationally Designed Self-assembling tetrahedron protein, T310 | | Descriptor: | GLYCEROL, Putative acetyltransferase SACOL2570 | | Authors: | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
4EDI
 
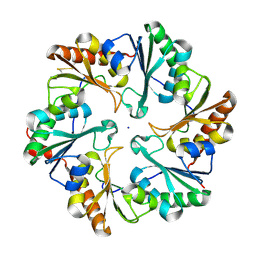 | | Disulfide bonded EutL from Clostridium perfringens | | Descriptor: | Ethanolamine utilization protein, SODIUM ION | | Authors: | Thompson, M.C, Cascio, D, Crowley, C.S, Kopstein, J.S, Yeates, T.O. | | Deposit date: | 2012-03-27 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | An allosteric model for control of pore opening by substrate binding in the EutL microcompartment shell protein.
Protein Sci., 24, 2015
|
|
4DCL
 
 | | Computationally Designed Self-assembling tetrahedron protein, T308, Crystallized in space group F23 | | Descriptor: | Putative acetyltransferase SACOL2570 | | Authors: | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2012-01-17 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
4FDZ
 
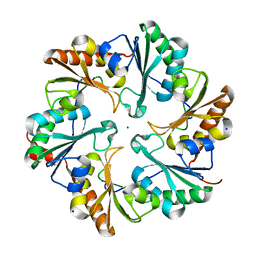 | | EutL from Clostridium perfringens, Crystallized Under Reducing Conditions | | Descriptor: | Ethanolamine utilization protein, SODIUM ION | | Authors: | Thompson, M.C, Cascio, D, Crowley, C.S, Kopstein, J.S, Yeates, T.O. | | Deposit date: | 2012-05-29 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | An allosteric model for control of pore opening by substrate binding in the EutL microcompartment shell protein.
Protein Sci., 24, 2015
|
|
1F1F
 
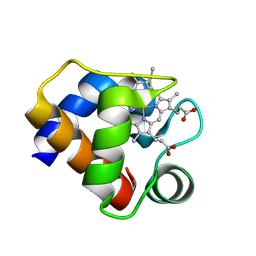 | | CRYSTAL STRUCTURE OF CYTOCHROME C6 FROM ARTHROSPIRA MAXIMA | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Serag, A.A, Sawaya, M.R, Krogmann, D.W, Yeates, T.O. | | Deposit date: | 2000-05-18 | | Release date: | 2001-08-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of cytochrome c-549 and cytochrome c6 from the cyanobacterium Arthrospira maxima.
Biochemistry, 40, 2001
|
|
1CYI
 
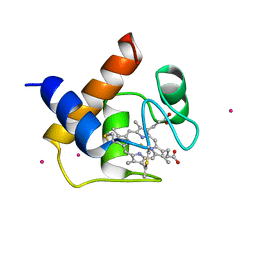 | | CYTOCHROME C6 | | Descriptor: | CADMIUM ION, CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 1995-05-09 | | Release date: | 1996-01-29 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of chloroplast cytochrome c6 at 1.9 A resolution: evidence for functional oligomerization.
J.Mol.Biol., 250, 1995
|
|
1CYJ
 
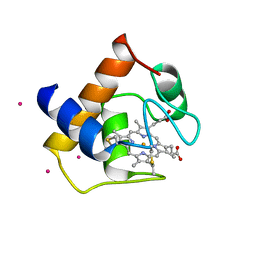 | | CYTOCHROME C6 | | Descriptor: | CADMIUM ION, CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 1995-05-09 | | Release date: | 1996-01-29 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of chloroplast cytochrome c6 at 1.9 A resolution: evidence for functional oligomerization.
J.Mol.Biol., 250, 1995
|
|
1DOF
 
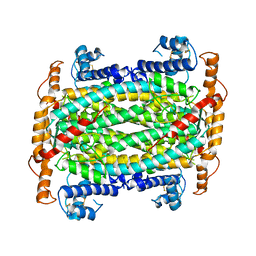 | | THE CRYSTAL STRUCTURE OF ADENYLOSUCCINATE LYASE FROM PYROBACULUM AEROPHILUM: INSIGHTS INTO THERMAL STABILITY AND HUMAN PATHOLOGY | | Descriptor: | ADENYLOSUCCINATE LYASE | | Authors: | Toth, E.A, Yeates, T.O, Goedken, E, Dixon, J.E, Marqusee, S. | | Deposit date: | 1999-12-20 | | Release date: | 2001-01-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of adenylosuccinate lyase from Pyrobaculum aerophilum reveals an intracellular protein with three disulfide bonds.
J.Mol.Biol., 301, 2000
|
|
4I7A
 
 | | GrpN pentameric microcompartment shell protein from Rhodospirillum rubrum | | Descriptor: | CHLORIDE ION, Ethanolamine utilization protein EutN/carboxysome structural protein Ccml | | Authors: | Wheatley, N.M, Gidaniyan, S.D, Cascio, D, Yeates, T.O. | | Deposit date: | 2012-11-30 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Bacterial microcompartment shells of diverse functional types possess pentameric vertex proteins.
Protein Sci., 22, 2013
|
|
4IVJ
 
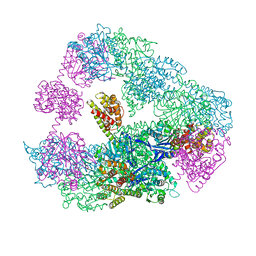 | | Structure of a 16 nm protein cage designed by fusing symmetric oligomeric domains, triple mutant, I222 form | | Descriptor: | Non-haem bromoperoxidase BPO-A2, Matrix protein 1 | | Authors: | Lai, Y.-T, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2013-01-23 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (7.351 Å) | | Cite: | Structure and flexibility of nanoscale protein cages designed by symmetric self-assembly.
J.Am.Chem.Soc., 135, 2013
|
|
4IQ4
 
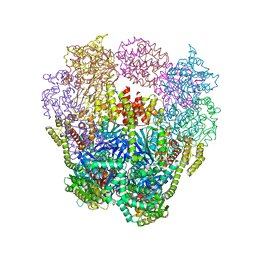 | | Structure of a 16 nm protein cage designed by fusing symmetric oligomeric domains, triple mutant, P21212 form | | Descriptor: | Non-haem bromoperoxidase BPO-A2, Matrix protein 1 | | Authors: | Lai, Y.-T, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2013-01-10 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.495 Å) | | Cite: | Structure and flexibility of nanoscale protein cages designed by symmetric self-assembly.
J.Am.Chem.Soc., 135, 2013
|
|
4ITV
 
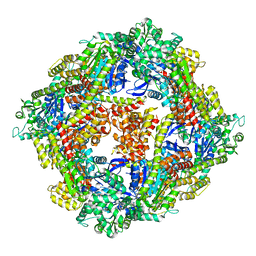 | | Structure of a 16 nm protein cage designed by fusing symmetric oligomeric domains, triple mutant, P212121 form | | Descriptor: | Non-haem bromoperoxidase BPO-A2, Matrix protein 1 | | Authors: | Lai, Y.-T, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2013-01-18 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.598 Å) | | Cite: | Structure and flexibility of nanoscale protein cages designed by symmetric self-assembly.
J.Am.Chem.Soc., 135, 2013
|
|
1F1C
 
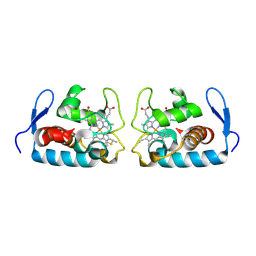 | | CRYSTAL STRUCTURE OF CYTOCHROME C549 | | Descriptor: | CYTOCHROME C549, HEME C | | Authors: | Kerfeld, C.A, Sawaya, M.R, Yeates, T.O, Krogmann, D.W. | | Deposit date: | 2000-05-18 | | Release date: | 2001-08-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of cytochrome c-549 and cytochrome c6 from the cyanobacterium Arthrospira maxima.
Biochemistry, 40, 2001
|
|
1JG1
 
 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with S-ADENOSYL-L-HOMOCYSTEINE | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, protein-L-isoaspartate O-methyltransferase | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
1JG4
 
 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with S-adenosylmethionine | | Descriptor: | S-ADENOSYLMETHIONINE, protein-L-isoaspartate O-methyltransferase | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
1L2H
 
 | | Crystal structure of Interleukin 1-beta F42W/W120F mutant | | Descriptor: | Interleukin 1-beta | | Authors: | Rudolph, M.G, Kelker, M.S, Schneider, T.R, Yeates, T.O, Oseroff, V, Heidary, D.K, Jennings, P.A, Wilson, I.A. | | Deposit date: | 2002-02-21 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Use of multiple anomalous dispersion to phase highly merohedrally twinned crystals of interleukin-1beta.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1KR5
 
 | | Crystal structure of human L-isoaspartyl methyltransferase | | Descriptor: | Protein-L-isoaspartate O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Ryttersgaard, C, Griffith, S.C, Sawaya, M.R, MacLaren, D.C, Clarke, S, Yeates, T.O. | | Deposit date: | 2002-01-08 | | Release date: | 2002-02-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human L-isoaspartyl methyltransferase.
J.Biol.Chem., 277, 2002
|
|
1JG3
 
 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with adenosine & VYP(ISP)HA substrate | | Descriptor: | ADENOSINE, CHLORIDE ION, SODIUM ION, ... | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2011-07-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
1JG2
 
 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with adenosine | | Descriptor: | ADENOSINE, SODIUM ION, protein-L-isoaspartate O-methyltransferase | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
8TFS
 
 | |
8T0B
 
 | |
8T1N
 
 | | Micro-ED Structure of a Novel Domain of Unknown Function Solved with AlphaFold | | Descriptor: | DUF1842 domain-containing protein | | Authors: | Miller, J.E, Cascio, D, Sawaya, M.R, Cannon, K.A, Rodriguez, J.A, Yeates, T.O. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | AlphaFold-assisted structure determination of a bacterial protein of unknown function using X-ray and electron crystallography.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8T1M
 
 | | Novel Domain of Unknown Function Solved with AlphaFold | | Descriptor: | DUF1842 domain-containing protein | | Authors: | Miller, J.E, Agdanowski, M.P, Cascio, D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | AlphaFold-assisted structure determination of a bacterial protein of unknown function using X-ray and electron crystallography.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8UMP
 
 | | T33-ml35 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms | | Descriptor: | T33-ml35-redesigned-CutA-fold, T33-ml35-redesigned-TPR-domain-fold | | Authors: | Castells-Graells, R, Meador, K, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-10-18 | | Release date: | 2023-11-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | A suite of designed protein cages using machine learning and protein fragment-based protocols.
Structure, 32, 2024
|
|
