3RIY
 
 | |
5TFP
 
 | |
8FRR
 
 | | Wild-type myocilin olfactomedin domain | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Ma, M.T, Lieberman, R.L, Huard, D.J.E. | | Deposit date: | 2023-01-08 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Competition between inside-out unfolding and pathogenic aggregation in an amyloid-forming beta-propeller.
Nat Commun, 15, 2024
|
|
8K8K
 
 | | Structure of Klebsiella pneumonia ModA | | Descriptor: | Molybdate transporter periplasmic protein | | Authors: | Zhao, Q, Bartlam, M. | | Deposit date: | 2023-07-31 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural analysis of molybdate binding protein ModA from Klebsiella pneumoniae.
Biochem.Biophys.Res.Commun., 681, 2023
|
|
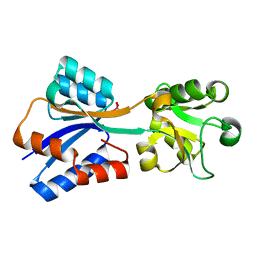
8K8L
 
 | |
7CMK
 
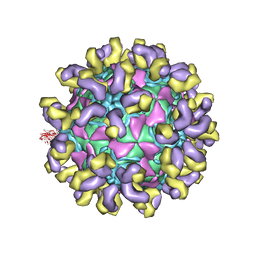 | | E30 E-particle in complex with 6C5 | | Descriptor: | Heavy chain, Light chain, VP1, ... | | Authors: | Wang, K, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-07-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Serotype specific epitopes identified by neutralizing antibodies underpin immunogenic differences in Enterovirus B.
Nat Commun, 11, 2020
|
|
8HE4
 
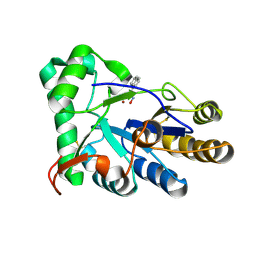 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION, ~{N}-oxidanylnaphthalene-1-carboxamide | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HE2
 
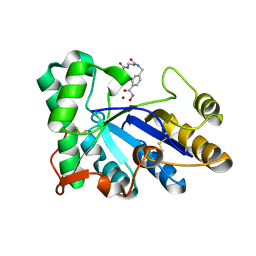 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION, tert-butyl N-[3-[[4-(oxidanylcarbamoyl)phenyl]methylamino]-3-oxidanylidene-propyl]carbamate | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HE1
 
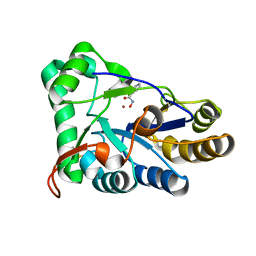 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | BENZHYDROXAMIC ACID, Chitin deacetylase, ZINC ION | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HFA
 
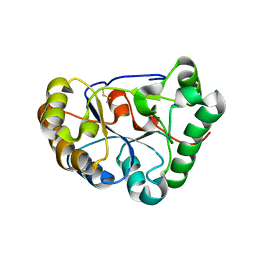 | |
8HF9
 
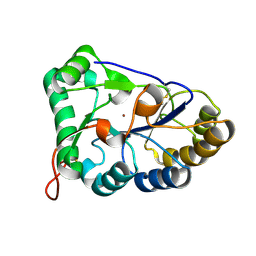 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-10 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HJF
 
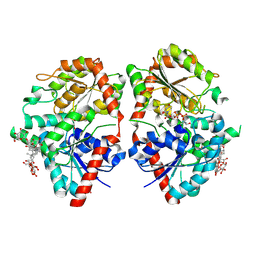 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with M5, state 2 | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, (2S,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(2R,3S,4S,5R,6R)-6-[[(3S,8S,9R,10R,11R,13R,14S,17R)-17-[(2S,5R)-5-[(2S,3R,4S,5S,6R)-3-[(2R,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-6-[[(2R,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxymethyl]-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-methyl-6-oxidanyl-heptan-2-yl]-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-3-yl]oxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methoxy]oxane-3,4,5-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJP
 
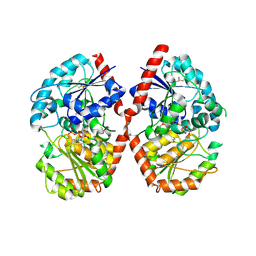 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with UDP state 1 | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, URIDINE-5'-DIPHOSPHATE, glycosyltranseferease | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJO
 
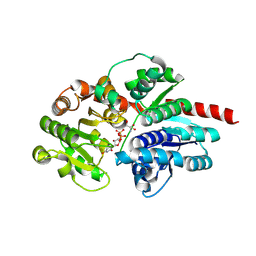 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with UDP state 2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJG
 
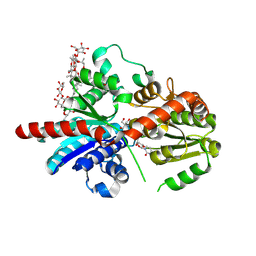 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with M5, state 1 | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, (2S,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(2R,3S,4S,5R,6R)-6-[[(3S,8S,9R,10R,11R,13R,14S,17R)-17-[(2S,5R)-5-[(2S,3R,4S,5S,6R)-3-[(2R,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-6-[[(2R,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxymethyl]-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-methyl-6-oxidanyl-heptan-2-yl]-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-3-yl]oxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methoxy]oxane-3,4,5-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJN
 
 | |
8HJQ
 
 | |
8HJH
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with SIA, state 3 | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, (2S,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(2S,3S,4S,5R,6S)-5-[(2S,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-6-[(3R,6S)-6-[(3S,8S,9R,10R,11S,13R,14S,17R)-3-[(2S,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-2-oxidanyl-heptan-3-yl]oxy-3,4-bis(oxidanyl)oxan-2-yl]methoxy]oxane-3,4,5-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJL
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with M3E | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, (2R,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(3S,8S,9R,10S,11S,13R,14S,17S)-17-[(2S,5R)-5-[(2S,3R,4S,5R,6S)-6-(hydroxymethyl)-3-[(2S,3R,4S,5S,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-methyl-6-oxidanyl-heptan-2-yl]-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-3-yl]oxy]oxane-3,4,5-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJK
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with SIA, state 1 | | Descriptor: | (2S,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(2S,3S,4S,5R,6S)-5-[(2S,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-6-[(3R,6S)-6-[(3S,8S,9R,10R,11S,13R,14S,17R)-3-[(2S,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-2-oxidanyl-heptan-3-yl]oxy-3,4-bis(oxidanyl)oxan-2-yl]methoxy]oxane-3,4,5-triol, URIDINE-5'-DIPHOSPHATE, glycosyltranseferease | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
6J0T
 
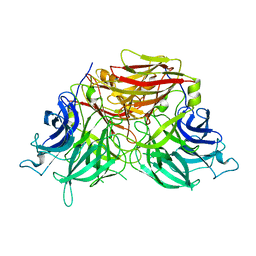 | | The crystal structure of exoinulinase INU1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Inulinase, MAGNESIUM ION | | Authors: | Hu, X.-J. | | Deposit date: | 2018-12-26 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure Study of Exoinulinase INU1
To Be Published
|
|
7EDO
 
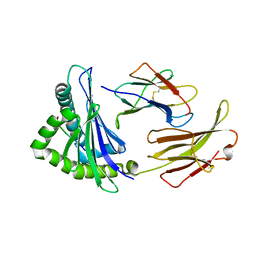 | | First insight into marsupial MHC I peptide presentation: immune features of lower mammals paralleled with bats | | Descriptor: | Beta-2-microglobulin, CYS-ASN-VAL-THR-LEU-ASN-TYR-PRO, MHC class I antigen | | Authors: | Wang, P.Y, Yue, C, Lu, D, Liu, K.F, Liu, S, Yao, S.J, Chai, Y, Qi, J.X, Lou, Y.L, Sun, Z.Y, Gao, G.F, Liu, W.J. | | Deposit date: | 2021-03-16 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Peptide Presentations of Marsupial MHC Class I Visualize Immune Features of Lower Mammals Paralleled with Bats.
J Immunol., 207, 2021
|
|
8X01
 
 | |
8YXR
 
 | |
8YXM
 
 | |
