1UFW
 
 | | Solution structure of RNP domain in Synaptojanin 2 | | Descriptor: | Synaptojanin 2 | | Authors: | He, F, Muto, Y, Ushikoshi, R, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nakayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-10 | | Release date: | 2003-12-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNP domain in Synaptojanin 2
To be Published
|
|
1UGK
 
 | | Solution structure of the first C2 domain of synaptotagmin IV from human fetal brain (KIAA1342) | | Descriptor: | Synaptotagmin IV | | Authors: | Nagashima, T, Hayashi, F, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nakayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-16 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first C2 domain of synaptotagmin IV from human fetal brain (KIAA1342)
To be Published
|
|
1UGV
 
 | | Solution structure of the SH3 domain of human olygophrein-1 like protein (KIAA0621) | | Descriptor: | Olygophrenin-1 like protein | | Authors: | Inoue, K, Hayashi, F, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nakayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-20 | | Release date: | 2003-12-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of human olygophrein-1 like protein (KIAA0621)
To be Published
|
|
1UG7
 
 | | Solution structure of four helical up-and-down bundle domain of the hypothetical protein 2610208M17Rik similar to the protein FLJ12806 | | Descriptor: | 2610208M17Rik protein | | Authors: | Li, H, Kigawa, T, Tomizawa, T, Koshiba, S, Inoue, M, Shirouzu, M, Terada, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-13 | | Release date: | 2004-08-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of four helical up-and-down bundle domain of the hypothetical protein 2610208M17Rik similar to the protein FLJ12806
To be Published
|
|
1UFX
 
 | | Solution structure of the third PDZ domain of human KIAA1526 protein | | Descriptor: | KIAA1526 protein | | Authors: | Tochio, N, Kobayashi, N, Koshiba, S, Kigawa, T, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nakayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-10 | | Release date: | 2003-12-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third PDZ domain of human KIAA1526 protein
To be Published
|
|
1UG1
 
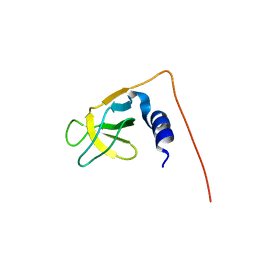 | | SH3 domain of Hypothetical protein BAA76854.1 | | Descriptor: | KIAA1010 protein | | Authors: | Nagata, T, Muto, Y, Kamewari, Y, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nakayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-11 | | Release date: | 2003-12-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of SH3 domain of Hypothetical protein BAA76854.1
To be Published
|
|
1UFN
 
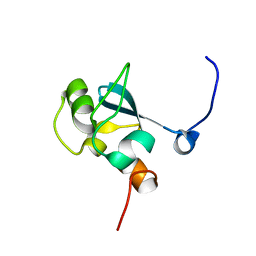 | | Solution structure of the SAND domain of the putative nuclear protein homolog (5830484A20Rik) | | Descriptor: | putative nuclear protein homolog 5830484A20Rik | | Authors: | Tochio, N, Kobayashi, N, Koshiba, S, Kigawa, T, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Matsuo, Y, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-02 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAND domain of the putative nuclear protein homolog (5830484A20Rik)
To be Published
|
|
1UFZ
 
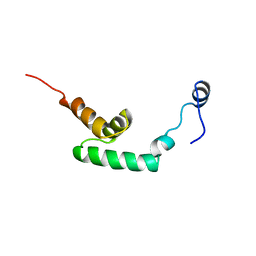 | | Solution structure of HBS1-like domain in hypothetical protein BAB28515 | | Descriptor: | Hypothetical protein BAB28515 | | Authors: | He, F, Muto, Y, Hayami, N, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-11 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of HBS1-like domain in hypothetical protein BAB28515
To be Published
|
|
1UG0
 
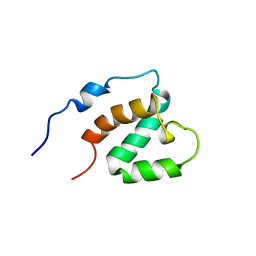 | | Solution structure of SURP domain in BAB30904 | | Descriptor: | splicing factor 4 | | Authors: | He, F, Muto, Y, Ushikoshi, R, Koshiba, S, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-11 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SURP domain in BAB30904
To be Published
|
|
1UGO
 
 | | Solution structure of the first Murine BAG domain of Bcl2-associated athanogene 5 | | Descriptor: | Bcl2-associated athanogene 5 | | Authors: | Endoh, H, Hayashi, F, Seimiya, K, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-17 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The C-terminal BAG domain of BAG5 induces conformational changes of the Hsp70 nucleotide-binding domain for ADP-ATP exchange
Structure, 18, 2010
|
|
1UGJ
 
 | | Solution structure of a murine hypothetical protein from RIKEN cDNA 2310057J16 | | Descriptor: | RIKEN cDNA 2310057J16 protein | | Authors: | Nagashima, T, Hayashi, F, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-16 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a murine hypothetical protein from RIKEN cDNA 2310057J16
To be Published
|
|
1UG8
 
 | | NMR structure of the R3H domain from Poly(A)-specific Ribonuclease | | Descriptor: | Poly(A)-specific Ribonuclease | | Authors: | Nagata, T, Muto, Y, Hayami, N, Uda, H, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-15 | | Release date: | 2004-08-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the R3H domain from Poly(A)-specific Ribonuclease
To be Published
|
|
1UDX
 
 | | Crystal structure of the conserved protein TT1381 from Thermus thermophilus HB8 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, the GTP-binding protein Obg | | Authors: | Kukimoto-Niino, M, Shirouzu, M, Murayama, K, Inoue, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-07 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of the GTP-binding protein Obg from Thermus thermophilus HB8.
J.Mol.Biol., 337, 2004
|
|
5GV0
 
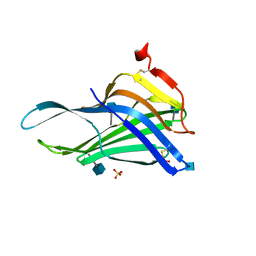 | | Crystal structure of the membrane-proximal domain of mouse lysosome-associated membrane protein 1 (LAMP-1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome-associated membrane glycoprotein 1, SULFATE ION | | Authors: | Tomabechi, Y, Ehara, H, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-09-01 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Lysosome-associated membrane proteins-1 and -2 (LAMP-1 and LAMP-2) assemble via distinct modes
Biochem.Biophys.Res.Commun., 479, 2016
|
|
5GV3
 
 | | Crystal structure of the membrane-distal domain of mouse lysosome-associated membrane protein 2 (LAMP-2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome-associated membrane glycoprotein 2, ZINC ION | | Authors: | Tomabechi, Y, Ehara, H, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-09-01 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Lysosome-associated membrane proteins-1 and -2 (LAMP-1 and LAMP-2) assemble via distinct modes.
Biochem. Biophys. Res. Commun., 479, 2016
|
|
1UG2
 
 | | Solution Structure of Mouse Hypothetical Gene (2610100B20Rik) Product Homologous to Myb DNA-binding Domain | | Descriptor: | 2610100B20Rik gene product | | Authors: | Zhao, C, Kigawa, T, Tochio, N, Koshiba, S, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-11 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Mouse Hypothetical Gene (2610100B20Rik) Product Homologous to Myb DNA-binding Domain
To be Published
|
|
5HNY
 
 | | Structural basis of backwards motion in kinesin-14: plus-end directed nKn669 in the AMPPNP state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
5HNZ
 
 | | Structural basis of backwards motion in kinesin-14: plus-end directed nKn669 in the nucleotide-free state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
8JHK
 
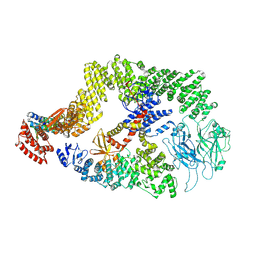 | | Cryo-EM structure of the DOCK5/ELMO1 complex, focused on one protomer | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.76 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
5HNX
 
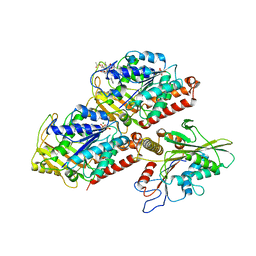 | | Structural basis of backwards motion in kinesin-14: minus-end directed nKn664 in the nucleotide-free state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
5HNW
 
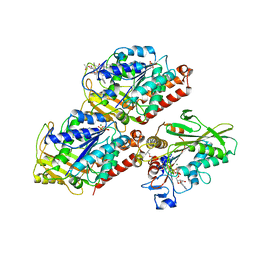 | | Structural basis of backwards motion in kinesin-14: minus-end directed nKn664 in the AMPPNP state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
8JG5
 
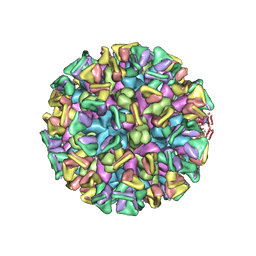 | | Cryo-EM structure of the GI.4 Chiba VLP complexed with the CV-1A1 Fv-clasp | | Descriptor: | VH,SARAH, VL,SARAH, VP1 | | Authors: | Hosaka, T, Katsura, K, Kimura-Someya, T, Someya, Y, Shirouzu, M. | | Deposit date: | 2023-05-19 | | Release date: | 2024-04-17 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural analyses of the GI.4 norovirus by cryo-electron microscopy and X-ray crystallography revealing binding sites for human monoclonal antibodies.
J.Virol., 98, 2024
|
|
5XON
 
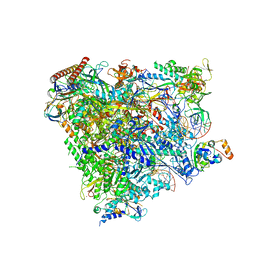 | | RNA Polymerase II elongation complex bound with Spt4/5 and TFIIS | | Descriptor: | DNA (48-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ehara, H, Yokoyama, T, Shigematsu, H, Shirouzu, M, Sekine, S. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Structure of the complete elongation complex of RNA polymerase II with basal factors
Science, 357, 2017
|
|
1UAN
 
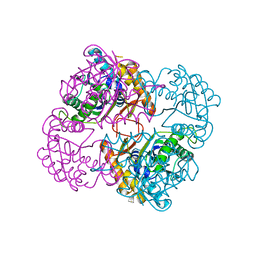 | | Crystal structure of the conserved protein TT1542 from Thermus thermophilus HB8 | | Descriptor: | hypothetical protein TT1542 | | Authors: | Handa, N, Terada, T, Tame, J.R.H, Park, S.-Y, Kinoshita, K, Ota, M, Nakamura, H, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-03-12 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the conserved protein TT1542 from Thermus thermophilus HB8
PROTEIN SCI., 12, 2003
|
|
1UC8
 
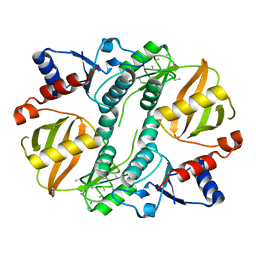 | | Crystal structure of a lysine biosynthesis enzyme, Lysx, from thermus thermophilus HB8 | | Descriptor: | lysine biosynthesis enzyme | | Authors: | Sakai, H, Vassylyeva, M.N, Matsuura, T, Sekine, S, Nishiyama, M, Terada, T, Shirouzu, M, Kuramitsu, S, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-04-09 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Lysine Biosynthesis Enzyme, LysX, from Thermus thermophilus HB8
J.Mol.Biol., 332, 2003
|
|
