3IYC
 
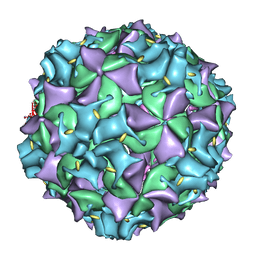 | | Poliovirus late RNA-release intermediate | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Genome polyprotein, ... | | Authors: | Levy, H.C, Bostina, M, Filman, D.J, Hogle, J.M. | | Deposit date: | 2009-07-21 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy.
J.Virol., 84, 2010
|
|
3JBE
 
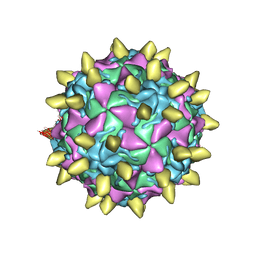 | | Complex of poliovirus with VHH PVSS8A | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
3J3P
 
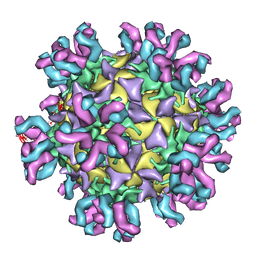 | | Conformational Shift of a Major Poliovirus Antigen Confirmed by Immuno-Cryogenic Electron Microscopy: 135S Poliovirus and C3-Fab Complex | | Descriptor: | C3 antibody, heavy chain, light chain, ... | | Authors: | Lin, J, Cheng, N, Hogle, J.M, Steven, A.C, Belnap, D.M. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Conformational shift of a major poliovirus antigen confirmed by immuno-cryogenic electron microscopy.
J.Immunol., 191, 2013
|
|
3J8F
 
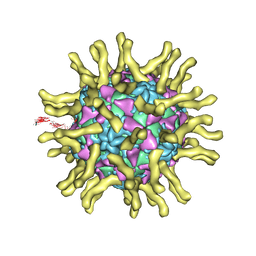 | | Cryo-EM reconstruction of poliovirus-receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, ... | | Authors: | Strauss, M, Filman, D.J, Belnap, D.M, Cheng, N, Noel, R.T, Hogle, J.M. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-11 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Nectin-Like Interactions between Poliovirus and Its Receptor Trigger Conformational Changes Associated with Cell Entry.
J.Virol., 89, 2015
|
|
3J48
 
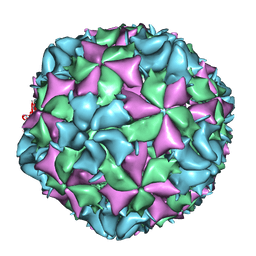 | | Cryo-EM structure of Poliovirus 135S particles | | Descriptor: | Protein VP1, Protein VP2, Protein VP3 | | Authors: | Butan, C, Fiman, D.J, Hogle, J.M. | | Deposit date: | 2013-06-28 | | Release date: | 2013-12-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Cryo-Electron Microscopy Reconstruction Shows Poliovirus 135S Particles Poised for Membrane Interaction and RNA Release.
J.Virol., 88, 2014
|
|
3J3O
 
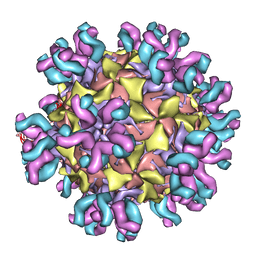 | | Conformational Shift of a Major Poliovirus Antigen Confirmed by Immuno-Cryogenic Electron Microscopy: 160S Poliovirus and C3-Fab Complex | | Descriptor: | C3 antibody, heavy chain, light chain, ... | | Authors: | Lin, J, Cheng, N, Hogle, J.M, Steven, A.C, Belnap, D.M. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.1 Å) | | Cite: | Conformational shift of a major poliovirus antigen confirmed by immuno-cryogenic electron microscopy.
J.Immunol., 191, 2013
|
|
3JBF
 
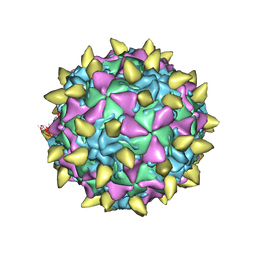 | | Complex of poliovirus with VHH PVSP19B | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
3JBD
 
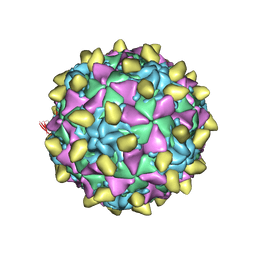 | | Complex of poliovirus with VHH PVSP6A | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Strauss, M, Schotte, L, Thys, B, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-27 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Five of Five VHHs Neutralizing Poliovirus Bind the Receptor-Binding Site.
J.Virol., 90, 2016
|
|
3ZX8
 
 | | Cryo-EM reconstruction of native and expanded Turnip Crinkle virus | | Descriptor: | CAPSID PROTEIN | | Authors: | Bakker, S.E, Robottom, J, Hogle, J.M, Maeda, A, Pearson, A.R, Stockley, P.G, Ranson, N.A, Harrison, S.C. | | Deposit date: | 2011-08-08 | | Release date: | 2012-07-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Isolation of an Asymmetric RNA Uncoating Intermediate for a Single-Stranded RNA Plant Virus.
J.Mol.Biol., 417, 2012
|
|
1ZC8
 
 | | Coordinates of tmRNA, SmpB, EF-Tu and h44 fitted into Cryo-EM map of the 70S ribosome and tmRNA complex | | Descriptor: | Elongation factor Tu, H2 16S rRNA, H2b d mRNA, ... | | Authors: | Valle, M, Gillet, R, Kaur, S, Henne, A, Ramakrishnan, V, Frank, J. | | Deposit date: | 2005-04-11 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Visualizing tmRNA Entry into a Stalled Ribosome
Science, 300, 2003
|
|
6RJF
 
 | | Echovirus 1 intact particle | | Descriptor: | PALMITIC ACID, VP1, VP2, ... | | Authors: | Domanska, A, Ruokolainen, V.P, Pelliccia, M, Laajala, M.A, Marjomaki, V.S, Butcher, S.J. | | Deposit date: | 2019-04-26 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Extracellular Albumin and Endosomal Ions Prime Enterovirus Particles for Uncoating That Can Be Prevented by Fatty Acid Saturation.
J.Virol., 93, 2019
|
|
1EAH
 
 | | PV2L COMPLEXED WITH ANTIVIRAL AGENT SCH48973 | | Descriptor: | 1[2-CHLORO-4-METHOXY-PHENYL-OXYMETHYL]-4-[2,6-DICHLORO-PHENYL-OXYMETHYL]-BENZENE, MYRISTIC ACID, POLIOVIRUS TYPE 2 COAT PROTEINS VP1 TO VP4 | | Authors: | Lentz, K, Arnold, E. | | Deposit date: | 1997-07-22 | | Release date: | 1998-09-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of poliovirus type 2 Lansing complexed with antiviral agent SCH48973: comparison of the structural and biological properties of three poliovirus serotypes.
Structure, 5, 1997
|
|
8EXX
 
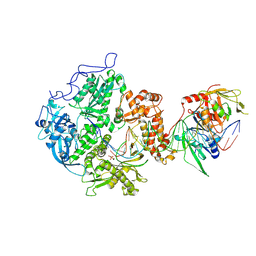 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA and foscarnet (pre-translocation state) | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2022-10-26 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
8V1T
 
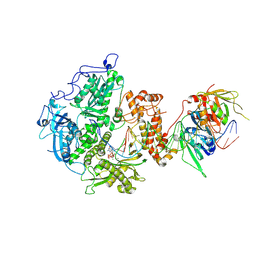 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA and acyclovir triphosphate in closed conformation | | Descriptor: | ACYCLOVIR TRIPHOSPHATE, DNA polymerase, DNA polymerase processivity factor, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
8V1R
 
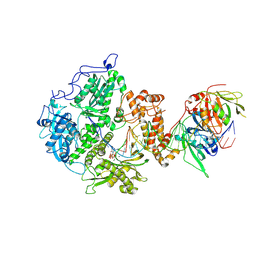 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA and DTTP in closed conformation | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
8V1Q
 
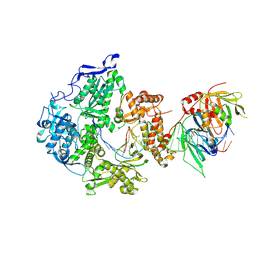 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA in both open/closed conformations | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
8V1S
 
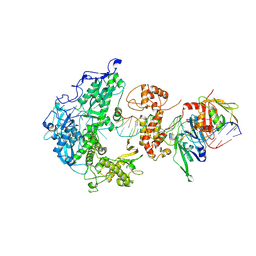 | | Herpes simplex virus 1 polymerase holoenzyme bound to mismatched DNA in editing conformation | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
2AIV
 
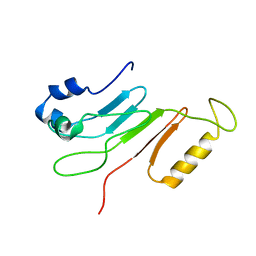 | | Multiple conformations in the ligand-binding site of the yeast nuclear pore targeting domain of NUP116P | | Descriptor: | fragment of Nucleoporin NUP116/NSP116 | | Authors: | Robinson, M.A, Park, S, Sun, Z.-Y.J, Silver, P, Wagner, G, Hogle, J. | | Deposit date: | 2005-08-01 | | Release date: | 2005-08-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Multiple Conformations in the Ligand-binding Site of the Yeast Nuclear Pore-targeting Domain of Nup116p
J.Biol.Chem., 280, 2005
|
|
2FJC
 
 | | Crystal structure of antigen TpF1 from Treponema pallidum | | Descriptor: | Antigen TpF1, FE (III) ION | | Authors: | Thumiger, A, Polenghi, A, Papinutto, E, Battistutta, R, Montecucco, C, Zanotti, G. | | Deposit date: | 2006-01-02 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of antigen TpF1 from Treponema pallidum.
Proteins, 62, 2006
|
|
2PYB
 
 | |
1JIG
 
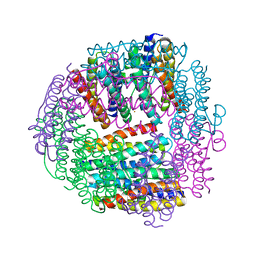 | | Dlp-2 from Bacillus anthracis | | Descriptor: | Dlp-2, FE (III) ION | | Authors: | Papinutto, E, Dundon, W.G, Pitulis, N, Battistutta, R, Montecucco, C, Zanotti, G. | | Deposit date: | 2001-07-02 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of two iron-binding proteins from Bacillus anthracis.
J.Biol.Chem., 277, 2002
|
|
3ZX9
 
 | | Cryo-EM reconstruction of native and expanded Turnip Crinkle virus | | Descriptor: | CAPSID PROTEIN | | Authors: | Bakker, S.E, Robottom, J, Pearson, A.R, Stockley, P.G, Ranson, N.A. | | Deposit date: | 2011-08-08 | | Release date: | 2012-07-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Isolation of an Asymmetric RNA Uncoating Intermediate for a Single-Stranded RNA Plant Virus.
J.Mol.Biol., 417, 2012
|
|
1JI4
 
 | | NAP protein from helicobacter pylori | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE (III) ION, NEUTROPHIL-ACTIVATING PROTEIN A, ... | | Authors: | Zanotti, G, Papinutto, E, Dundon, W.G, Battistutta, R, Seveso, M, Del Giudice, G, Rappuoli, R, Montecucco, C. | | Deposit date: | 2001-06-29 | | Release date: | 2002-10-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Neutrophil-activating Protein from Helicobacter pylori
J.Mol.Biol., 323, 2002
|
|
1JI5
 
 | | Dlp-1 from bacillus anthracis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Dlp-1, FE (III) ION | | Authors: | Papinutto, E, Dundon, W.G, Pitulis, N, Battistutta, R, Montecucco, C, Zanotti, G. | | Deposit date: | 2001-06-29 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of two iron-binding proteins from Bacillus anthracis.
J.Biol.Chem., 277, 2002
|
|
1O9R
 
 | | The X-ray crystal structure of Agrobacterium tumefaciens Dps, a member of the family that protect DNA without binding | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AGROBACTERIUM TUMEFACIENS DPS, ... | | Authors: | Ilari, A, Ceci, P, Chiancone, E. | | Deposit date: | 2002-12-18 | | Release date: | 2003-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Dps Protein of Agrobacterium Tumefaciens Does not Bind to DNA But Protects It Toward Oxidative Cleavage: X-Ray Crystal Structure, Iron Binding, and Hydroxyl-Radical Scavenging Properties
J.Biol.Chem., 278, 2003
|
|
