8Y4C
 
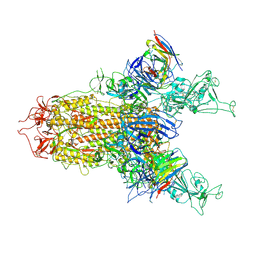 | | BA.2.86 S-trimer in complex with Nab XGv280 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, XGv280 Heavy chain, ... | | Authors: | Zhu, Q, Liu, P. | | Deposit date: | 2024-01-30 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Enhancing RBD exposure and S1 shedding by an extremely conserved SARS-CoV-2 NTD epitope.
Signal Transduct Target Ther, 9, 2024
|
|
8Y4A
 
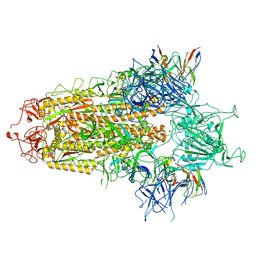 | | BA.2.86 S-trimer in complex with Nab XG2v046 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhu, Q, Liu, P. | | Deposit date: | 2024-01-30 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Enhancing RBD exposure and S1 shedding by an extremely conserved SARS-CoV-2 NTD epitope.
Signal Transduct Target Ther, 9, 2024
|
|
8W6P
 
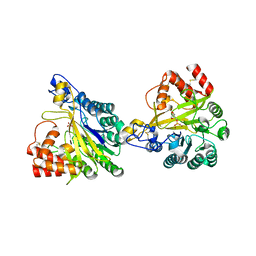 | | Crystal structure of dimeric murine SMPDL3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, ... | | Authors: | Zhang, C, Liu, P, Fan, S, Hou, Y. | | Deposit date: | 2023-08-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | SMPDL3A is a cGAMP-degrading enzyme induced by LXR-mediated lipid metabolism to restrict cGAS-STING DNA sensing.
Immunity, 56, 2023
|
|
8W6R
 
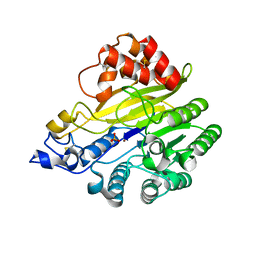 | | murine SMPDL3A bound to sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, ... | | Authors: | Zhang, C, Liu, P, Fan, S. | | Deposit date: | 2023-08-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SMPDL3A is a cGAMP-degrading enzyme induced by LXR-mediated lipid metabolism to restrict cGAS-STING DNA sensing.
Immunity, 56, 2023
|
|
6JHT
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F9 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6JHQ
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F4 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-18 | | Release date: | 2020-03-18 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6JHS
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F7 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6JHR
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F6 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-18 | | Release date: | 2020-03-18 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
8HBJ
 
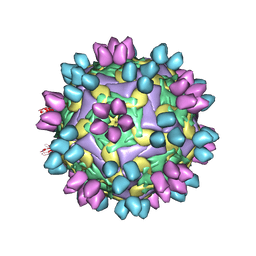 | | cocktail of FMDV (A/TUR/14/98) in complex with M678F and M688F | | Descriptor: | M678F nab, M688F nab, VP1 of capsid protein, ... | | Authors: | Li, H.Z, Dong, H, Liu, P. | | Deposit date: | 2022-10-28 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Foot-and-mouth disease virus antigenic landscape and reduced immunogenicity elucidated in atomic detail.
Nat Commun, 15, 2024
|
|
8HBG
 
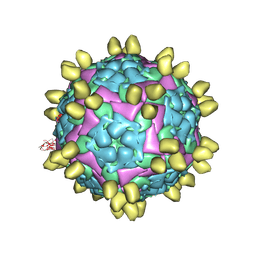 | | FMDV (A/TUR/14/98) in complex with M678F | | Descriptor: | M678F nab, VP1 of capsid protein, VP2 of capsid protein, ... | | Authors: | Li, H.Z, Dong, H, Liu, P. | | Deposit date: | 2022-10-28 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Foot-and-mouth disease virus antigenic landscape and reduced immunogenicity elucidated in atomic detail.
Nat Commun, 15, 2024
|
|
8HEG
 
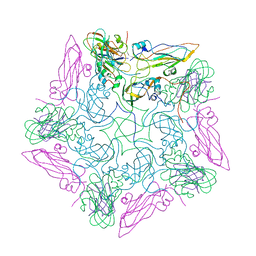 | | Pentamer of FMDV (A/TUR/14/98) in complex with M3F | | Descriptor: | M3F, VP1 of capsid protein, VP2 of capsid protein, ... | | Authors: | Li, H.Z, Dong, H, Liu, P. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-15 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Foot-and-mouth disease virus antigenic landscape and reduced immunogenicity elucidated in atomic detail.
Nat Commun, 15, 2024
|
|
8HBI
 
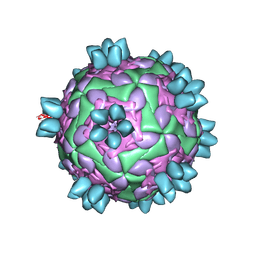 | | FMDV (A/TUR/14/98) in complex with M688F | | Descriptor: | M688F nab, VP1 of capsid protein, VP2 of capsid protein, ... | | Authors: | Li, H.Z, Dong, H, Liu, P. | | Deposit date: | 2022-10-28 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Foot-and-mouth disease virus antigenic landscape and reduced immunogenicity elucidated in atomic detail.
Nat Commun, 15, 2024
|
|
5ZQZ
 
 | | Structure of human mitochondrial trifunctional protein, tetramer | | Descriptor: | Trifunctional enzyme subunit alpha, mitochondrial, Trifunctional enzyme subunit beta | | Authors: | Liang, K, Li, N, Dai, J, Wang, X, Liu, P, Chen, X, Wang, C, Gao, N, Xiao, J. | | Deposit date: | 2018-04-20 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of human mitochondrial trifunctional protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5ZRV
 
 | | Structure of human mitochondrial trifunctional protein, octamer | | Descriptor: | Trifunctional enzyme subunit alpha, mitochondrial, Trifunctional enzyme subunit beta | | Authors: | Liang, K, Li, N, Dai, J, Wang, X, Liu, P, Chen, X, Wang, C, Gao, N, Xiao, J. | | Deposit date: | 2018-04-25 | | Release date: | 2018-06-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Cryo-EM structure of human mitochondrial trifunctional protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8Y32
 
 | | BA.2.86-V483 RBD in complex with bACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, CHLORIDE ION, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2024-01-28 | | Release date: | 2024-11-13 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | BA.2.86-V483 RBD in complex with bACE2
To Be Published
|
|
8XZ8
 
 | | BA.2.86 Spike in complex with bovine ACE2 (bound 1 ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, CHLORIDE ION, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2024-01-21 | | Release date: | 2024-11-13 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | BA.2.86 Spike in complex with bovine ACE2
To Be Published
|
|
8XZA
 
 | |
8XZ9
 
 | | BA.2.86 Spike in complex with bovine ACE2 (bound 2 ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2024-01-21 | | Release date: | 2024-11-13 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | BA.2.86 Spike in complex with bovine ACE2
To Be Published
|
|
7DST
 
 | | FMDV capsid in complex with M170 Nab | | Descriptor: | M170 Nab, VP1 of O type FMDV capsid, VP2 of O type FMDV capsid, ... | | Authors: | Dong, H, Liu, P. | | Deposit date: | 2021-01-02 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and molecular basis for foot-and-mouth disease virus neutralization by two potent protective antibodies.
Protein Cell, 13, 2022
|
|
7DSS
 
 | | Complex of FMDV and M8 Nab | | Descriptor: | M8 Nab, VP1 of O type FMDV capsid, VP2 of O-type FMDV capsid, ... | | Authors: | Dong, H, Liu, P. | | Deposit date: | 2021-01-02 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural and molecular basis for foot-and-mouth disease virus neutralization by two potent protective antibodies.
Protein Cell, 13, 2022
|
|
1N46
 
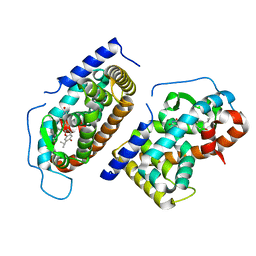 | | CRYSTAL STRUCTURE OF HUMAN TR BETA LIGAND-BINDING DOMAIN COMPLEXED WITH A POTENT SUBTYPE-SELECTIVE THYROMIMETIC | | Descriptor: | Thyroid hormone receptor Beta-1, [4-(4-HYDROXY-3-ISOPROPYL-PHENOXY)-3,5-DIMETHYL-PHENYL]-6-AZAURACIL | | Authors: | Dow, R.L, Schneider, S.R, Paight, E.S, Hank, R.F, Chiang, P, Cornelius, P, Lee, E, Newsome, W.P, Swick, A.G, Spitzer, J, Hargrove, D.M, Patterson, T.A, Pandit, J, Chrunyk, B.A, LeMotte, P.K, Danley, D.E, Rosner, M.H, Ammirati, M.J, Simons, S.P, Schulte, G.K, Tate, B.F, DaSilva-Jardine, P. | | Deposit date: | 2002-10-30 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Novel Series of 6-Azauracil-Based Thyroid Hormone Receptor Ligands:
Potent, TRbeta Subtype-Selective Thyromimetics
Bioorg.Med.Chem.Lett., 13, 2003
|
|
6TF9
 
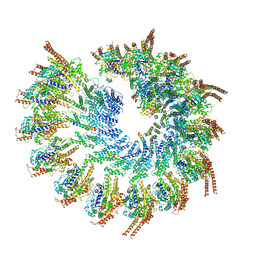 | | Structure of the vertebrate gamma-Tubulin Ring Complex | | Descriptor: | Actin, cytoplasmic 1, Belt helices 1,2,3,4, ... | | Authors: | Zupa, E, Pfeffer, S. | | Deposit date: | 2019-11-13 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Insights into the assembly and activation of the microtubule nucleator gamma-TuRC.
Nature, 578, 2020
|
|
9C45
 
 | |
9C44
 
 | |
7VHH
 
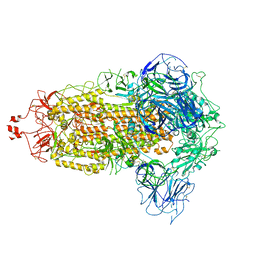 | | Delta variant of SARS-CoV-2 Spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Liu, P. | | Deposit date: | 2021-09-22 | | Release date: | 2022-12-28 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of delta variant SARS-CoV-2 Spike protein
To Be Published
|
|
