3MBX
 
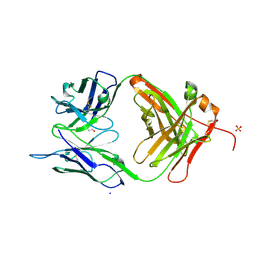 | | Crystal structure of chimeric antibody X836 | | Descriptor: | PHOSPHATE ION, SODIUM ION, X836 HEAVY CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L. | | Deposit date: | 2010-03-26 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | On the domain pairing in chimeric antibodies.
Mol.Immunol., 47, 2010
|
|
3MCK
 
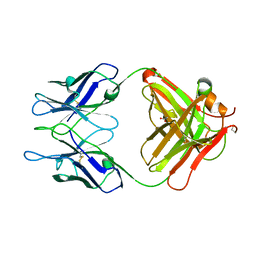 | |
3MCL
 
 | |
3O11
 
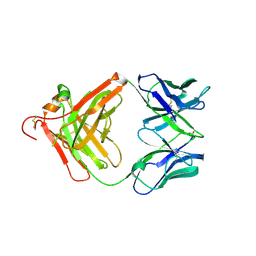 | | Anti-beta-amyloid antibody c706 fab in space group c2 | | Descriptor: | C706 HEAVY CHAIN variable region, Ig gamma-1 chain C region chimera, C706 LIGHT CHAIN variable region, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2010-07-20 | | Release date: | 2010-09-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | His-tag binding by antibody C706 mimics beta-amyloid recognition.
J.Mol.Recognit., 24, 2011
|
|
1SUB
 
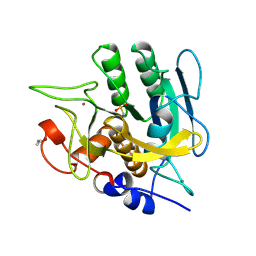 | | CALCIUM-INDEPENDENT SUBTILISIN BY DESIGN | | Descriptor: | ACETONE, CALCIUM ION, POTASSIUM ION, ... | | Authors: | Gallagher, T, Bryan, P, Gilliland, G.L. | | Deposit date: | 1992-06-10 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Calcium-independent subtilisin by design.
Proteins, 16, 1993
|
|
1SUA
 
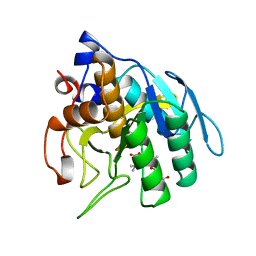 | | SUBTILISIN BPN' | | Descriptor: | SUBTILISIN BPN', TETRAPEPTIDE ALA-LEU-ALA-LEU | | Authors: | Almog, O, Gilliland, G.L. | | Deposit date: | 1997-01-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of calcium-independent subtilisin BPN' with restored thermal stability folded without the prodomain.
Proteins, 31, 1998
|
|
1SUE
 
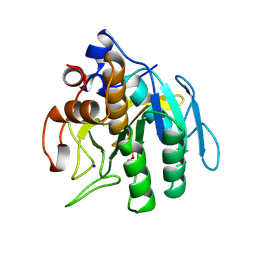 | | SUBTILISIN BPN' FROM BACILLUS AMYLOLIQUEFACIENS, MUTANT | | Descriptor: | DIISOPROPYL PHOSPHONATE, SODIUM ION, SUBTILISIN BPN' | | Authors: | Gallagher, D.T, Bryan, P, Pan, Q, Gilliland, G.L. | | Deposit date: | 1998-02-17 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of ionic strength dependence of crystal growth rates in a subtilisin variant.
J.Cryst.Growth, 193, 1998
|
|
1SUC
 
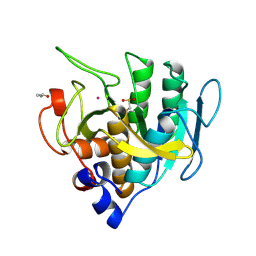 | |
1R4B
 
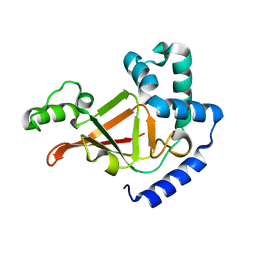 | |
1S4C
 
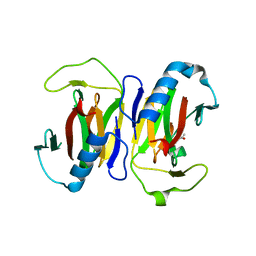 | | YHCH PROTEIN (HI0227) COPPER COMPLEX | | Descriptor: | ACETATE ION, COPPER (II) ION, Protein HI0227 | | Authors: | Teplyakov, A, Obmolova, G, Toedt, J, Gilliland, G.L. | | Deposit date: | 2004-01-15 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the bacterial YhcH protein indicates a role in sialic acid catabolism.
J.Bacteriol., 187, 2005
|
|
1R4W
 
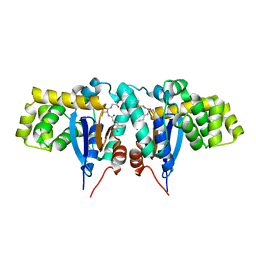 | | Crystal structure of Mitochondrial class kappa glutathione transferase | | Descriptor: | GLUTATHIONE, Glutathione S-transferase, mitochondrial | | Authors: | Ladner, J.E, Parsons, J.F, Rife, C.L, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2003-10-08 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Parallel Evolutionary Pathways for Glutathione Transferases: Structure and Mechanism of the Mitochondrial Class Kappa Enzyme rGSTK1-1
Biochemistry, 43, 2004
|
|
4KUZ
 
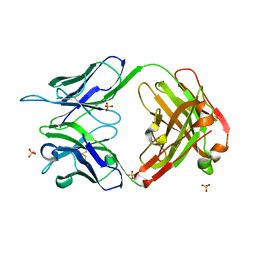 | |
1R45
 
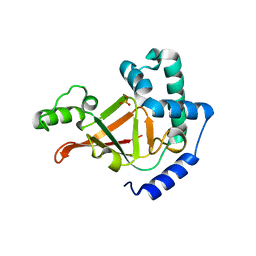 | | ADP-ribosyltransferase C3bot2 from Clostridium botulinum, triclinic form | | Descriptor: | GLYCEROL, Mono-ADP-ribosyltransferase C3, SULFATE ION | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L, Narumiya, S. | | Deposit date: | 2003-10-03 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of ADP-ribosyltransferase C3bot2 from Clostridium botulinum
To be Published
|
|
1RW1
 
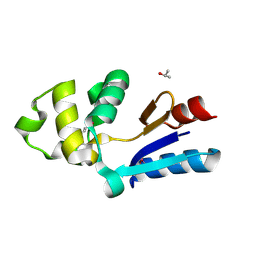 | | YFFB (PA3664) PROTEIN | | Descriptor: | ISOPROPYL ALCOHOL, conserved hypothetical protein yffB | | Authors: | Teplyakov, A, Pullalarevu, S, Obmolova, G, Doseeva, V, Galkin, A, Herzberg, O, Dauter, M, Dauter, Z, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2003-12-15 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structure of the YffB protein from Pseudomonas aeruginosa suggests a glutathione-dependent thiol reductase function.
Bmc Struct.Biol., 4, 2004
|
|
4KQ3
 
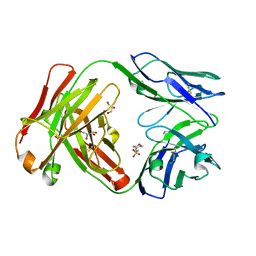 | | Crystal structure of Anti-IL-17A antibody CNTO3186 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CNTO3186 heavy chain, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-05-14 | | Release date: | 2014-03-26 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural evidence for a constrained conformation of short CDR-L3 in antibodies.
Proteins, 82, 2014
|
|
4LPW
 
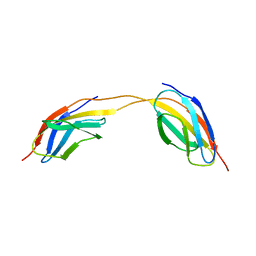 | |
4M7L
 
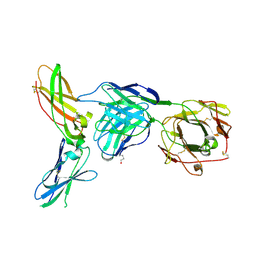 | |
4LPY
 
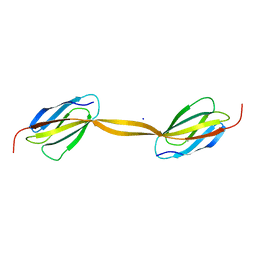 | |
1SUP
 
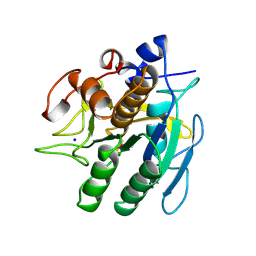 | | SUBTILISIN BPN' AT 1.6 ANGSTROMS RESOLUTION: ANALYSIS OF DISCRETE DISORDER AND COMPARISON OF CRYSTAL FORMS | | Descriptor: | CALCIUM ION, SODIUM ION, SUBTILISIN BPN', ... | | Authors: | Gallagher, D.T, Oliver, J.D, Betzel, C, Gilliland, G.L. | | Deposit date: | 1995-08-14 | | Release date: | 1995-11-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Subtilisin BPN' at 1.6 A resolution: analysis for discrete disorder and comparison of crystal forms.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
4LPU
 
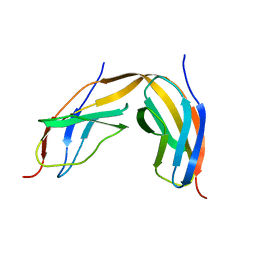 | |
4LPV
 
 | | Crystal structure of TENCON variant P41BR3-42 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TENCON variant P41BR3-42 | | Authors: | Teplyakov, A, Obmolova, G, Luo, J, Gilliland, G.L. | | Deposit date: | 2013-07-16 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C-terminal beta-strand swapping in a consensus-derived fibronectin Type III scaffold.
Proteins, 82, 2014
|
|
1SUD
 
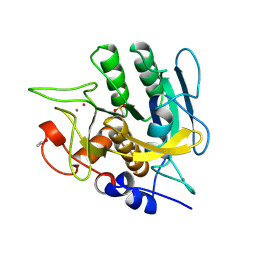 | | CALCIUM-INDEPENDENT SUBTILISIN BY DESIGN | | Descriptor: | ACETONE, CALCIUM ION, POTASSIUM ION, ... | | Authors: | Gallagher, T, Bryan, P, Gilliland, G.L. | | Deposit date: | 1992-06-10 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Calcium-independent subtilisin by design.
Proteins, 16, 1993
|
|
4KMT
 
 | | Crystal structure of human germline antibody 5-51/O12 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-05-08 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
4KQ4
 
 | | Crystal structure of Anti-IL-17A antibody CNTO7357 | | Descriptor: | CNTO7357 heavy chain, CNTO7357 light chain, NICKEL (II) ION | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-05-14 | | Release date: | 2014-03-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
4M6N
 
 | |
