6OZY
 
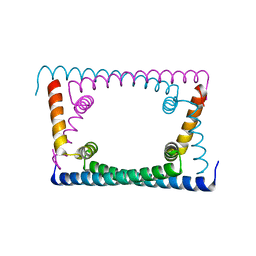 | | Wild type GapR crystal structure 2 from C. crescentus | | Descriptor: | CADMIUM ION, UPF0335 protein CC_3319 | | Authors: | Tarry, M, Harmel, C, Taylor, J.A, Marczynski, G.T, Schmeing, T.M. | | Deposit date: | 2019-05-16 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.014 Å) | | Cite: | Structures of GapR reveal a central channel which could accommodate B-DNA.
Sci Rep, 9, 2019
|
|
8V8M
 
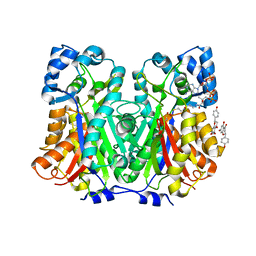 | | Switchgrass Chalcone Synthase | | Descriptor: | COENZYME A, Chalcone synthase, NARINGENIN | | Authors: | Lewis, J.A, Kang, C. | | Deposit date: | 2023-12-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural and Interactional Analysis of the Flavonoid Pathway Proteins: Chalcone Synthase, Chalcone Isomerase and Chalcone Isomerase-like Protein.
Int J Mol Sci, 25, 2024
|
|
8V8O
 
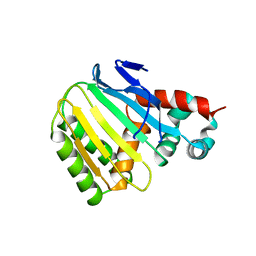 | | Switchgrass Chalcone Isomerase-Like Protein | | Descriptor: | Chalcone-flavonone isomerase family protein | | Authors: | Lewis, J.A, Kang, C. | | Deposit date: | 2023-12-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural and Interactional Analysis of the Flavonoid Pathway Proteins: Chalcone Synthase, Chalcone Isomerase and Chalcone Isomerase-like Protein.
Int J Mol Sci, 25, 2024
|
|
6CL8
 
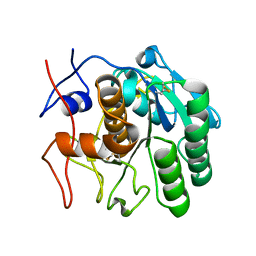 | | 2.00 A MicroED structure of proteinase K at 2.6 e- / A^2 | | Descriptor: | Proteinase K | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | ELECTRON CRYSTALLOGRAPHY (2 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLI
 
 | | 1.01 A MicroED structure of GSNQNNF at 0.17 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.01 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLR
 
 | | 1.31 A MicroED structure of GSNQNNF at 3.1 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.31 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
8V8P
 
 | | Sorghum Chalcone Isomerase | | Descriptor: | 7-HYDROXY-2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, Chalcone-flavonone isomerase family protein | | Authors: | Lewis, J.A, Kang, C. | | Deposit date: | 2023-12-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structural and Interactional Analysis of the Flavonoid Pathway Proteins: Chalcone Synthase, Chalcone Isomerase and Chalcone Isomerase-like Protein.
Int J Mol Sci, 25, 2024
|
|
8V8N
 
 | | Switchgrass Chalcone Synthase C170S | | Descriptor: | Chalcone synthase, GLYCEROL | | Authors: | Lewis, J.A, Kang, C. | | Deposit date: | 2023-12-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural and Interactional Analysis of the Flavonoid Pathway Proteins: Chalcone Synthase, Chalcone Isomerase and Chalcone Isomerase-like Protein.
Int J Mol Sci, 25, 2024
|
|
8V8L
 
 | | Switchgrass Chalcone Isomerase | | Descriptor: | Chalcone-flavonone isomerase family protein, GLYCEROL | | Authors: | Lewis, J.A, Kang, C. | | Deposit date: | 2023-12-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Interactional Analysis of the Flavonoid Pathway Proteins: Chalcone Synthase, Chalcone Isomerase and Chalcone Isomerase-like Protein.
Int J Mol Sci, 25, 2024
|
|
6CLF
 
 | | 1.15 A MicroED structure of GSNQNNF at 1.9 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.15 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLO
 
 | | 1.15 A MicroED structure of GSNQNNF at 2.1 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.15 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6PRY
 
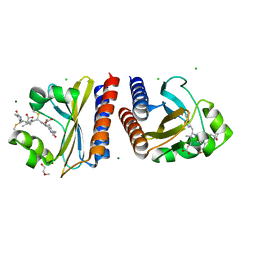 | | X-ray crystal structure of the blue-light absorbing state of PixJ from Thermosynechococcus elongatus by serial femtosecond crystallographic analysis | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Burgie, E.S, Clinger, J.A, Miller, M.D, Phillips Jr, G.N, Vierstra, R.D, Orville, A.M, Kern, J.F. | | Deposit date: | 2019-07-12 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P7M
 
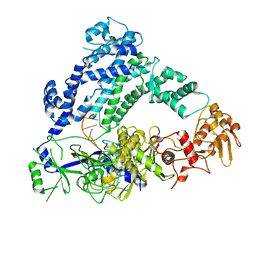 | | Cryo-EM structure of LbCas12a-crRNA: AcrVA4 (1:2 complex) | | Descriptor: | Cas12a, MAGNESIUM ION, anti-CRISPR VA4, ... | | Authors: | Knott, G.J, Liu, J.J, Doudna, J.A. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for AcrVA4 inhibition of specific CRISPR-Cas12a.
Elife, 8, 2019
|
|
7AJZ
 
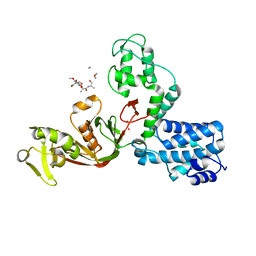 | | The X-ray Structure of L,D-transpeptidase LdtA from Vibrio cholerae in complex with NAG-NAM(tetrapeptide) | | Descriptor: | 1,2-ETHANEDIOL, L,D-transpeptidase YcbB, NAG-NAM(tetrapeptide), ... | | Authors: | Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | The X-ray Structure of L,D-transpeptidase LdtA from Vibrio cholerae in complex with NAG-NAM(tetrapeptide)
To Be Published
|
|
7AGZ
 
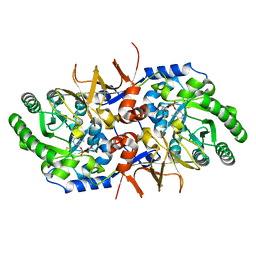 | | BsrV no-histagged | | Descriptor: | Broad specificity amino-acid racemase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Espaillat, A, Cava, F, Hermoso, J.A. | | Deposit date: | 2020-09-23 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Binding of non-canonical peptidoglycan controls Vibrio cholerae broad spectrum racemase activity.
Comput Struct Biotechnol J, 19, 2021
|
|
6CL9
 
 | | 2.20 A MicroED structure of proteinase K at 4.3 e- / A^2 | | Descriptor: | Proteinase K | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.2 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
7AJO
 
 | | The X-ray Structure of L,D-transpeptidase LdtA from Vibrio cholerae in complex with the cross-linking reaction intermediate | | Descriptor: | (2~{S},6~{S})-2-azanyl-6-[[(4~{R})-4-azanyl-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]heptanedioic acid, 1,2-ETHANEDIOL, L,D-transpeptidase YcbB | | Authors: | Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The X-ray Structure of L,D-transpeptidase LdtA from Vibrio cholerae in complex with the cross-linking reaction intermediate
To Be Published
|
|
6CLA
 
 | | 2.80 A MicroED structure of proteinase K at 6.0 e- / A^2 | | Descriptor: | Proteinase K | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
7AJX
 
 | | The X-ray Structure of L,D-transpeptidase LdtA from Vibrio cholerae in complex with meropenem | | Descriptor: | (2S,3R,4S)-4-{[(3S,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, L,D-transpeptidase YcbB | | Authors: | Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The X-ray Structure of L,D-transpeptidase LdtA from Vibrio cholerae in complex with meropenem
To Be Published
|
|
6CLG
 
 | | 1.35 A MicroED structure of GSNQNNF at 2.4 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.35 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLJ
 
 | | 1.01 A MicroED structure of GSNQNNF at 0.50 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.01 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLN
 
 | | 1.15 A MicroED structure of GSNQNNF at 1.8 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.15 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLQ
 
 | | 1.21 A MicroED structure of GSNQNNF at 2.8 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.21 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
7AJ9
 
 | |
6CFH
 
 | |
