4K1N
 
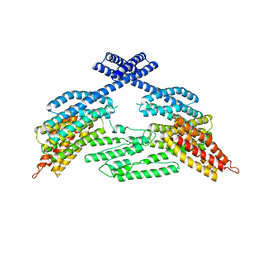 | |
8H2U
 
 | | X-ray Structure of photosystem I-LHCI super complex from Chlamydomonas reinhardtii. | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Tanaka, H, Kubota-Kawai, H, Misumi, Y, Kurisu, G. | | Deposit date: | 2022-10-07 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Three structures of PSI-LHCI from Chlamydomonas reinhardtii suggest a resting state re-activated by ferredoxin.
Biochim Biophys Acta Bioenerg, 1864, 2023
|
|
7V5N
 
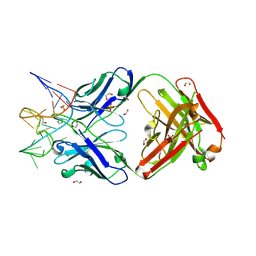 | | Crystal structure of Fab fragment of bevacizumab bound to DNA aptamer | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*GP*TP*TP*GP*GP*TP*GP*GP*TP*AP*GP*TP*TP*AP*CP*GP*TP*TP*CP*GP*C)-3'), IMIDAZOLE, ... | | Authors: | Hishiki, A, Tong, J, Todoroki, K, Hashimoto, H. | | Deposit date: | 2021-08-17 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of a DNA aptamer that binds to the complementarity-determining region of therapeutic monoclonal antibody and affinity improvement induced by pH-change for sensitive detection.
Biosens.Bioelectron., 203, 2022
|
|
6KX8
 
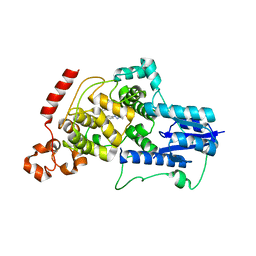 | | Crystal structure of mouse Cryptochrome 2 in complex with TH301 compound | | Descriptor: | 1-(4-chlorophenyl)-N-[2-(4-methoxyphenyl)-5,5-bis(oxidanylidene)-4,6-dihydrothieno[3,4-c]pyrazol-3-yl]cyclopentane-1-carboxamide, Cryptochrome-2 | | Authors: | Miller, S.A, Aikawa, Y, Hirota, T. | | Deposit date: | 2019-09-10 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Isoform-selective regulation of mammalian cryptochromes.
Nat.Chem.Biol., 16, 2020
|
|
6KX7
 
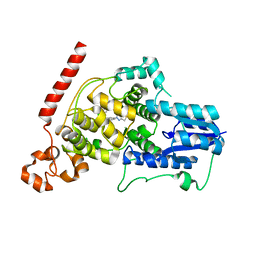 | | Crystal structure of mouse Cryptochrome 1 in complex with TH301 compound | | Descriptor: | 1-(4-chlorophenyl)-N-[2-(4-methoxyphenyl)-5,5-bis(oxidanylidene)-4,6-dihydrothieno[3,4-c]pyrazol-3-yl]cyclopentane-1-carboxamide, Cryptochrome-1 | | Authors: | Miller, S.A, Aikawa, Y, Hirota, T. | | Deposit date: | 2019-09-10 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Isoform-selective regulation of mammalian cryptochromes.
Nat.Chem.Biol., 16, 2020
|
|
6KX5
 
 | |
6KX6
 
 | | Crystal structure of mouse Cryptochrome 1 in complex with KL101 compound | | Descriptor: | Cryptochrome-1, ~{N}-[2-(2,4-dimethylphenyl)-4,6-dihydrothieno[3,4-c]pyrazol-3-yl]-3,4-dimethyl-benzamide | | Authors: | Miller, S.A, Aikawa, Y, Hirota, T. | | Deposit date: | 2019-09-10 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Isoform-selective regulation of mammalian cryptochromes.
Nat.Chem.Biol., 16, 2020
|
|
6KX4
 
 | |
7WYI
 
 | | Native Photosystem I of Chlamydomonas reinhardtii | | Descriptor: | CHLOROPHYLL A, CHLOROPHYLL A ISOMER, CHLOROPHYLL B, ... | | Authors: | Kurisu, G, Gerle, C, Mitsuoka, K, Kawamoto, A, Tanaka, H. | | Deposit date: | 2022-02-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Three structures of PSI-LHCI from Chlamydomonas reinhardtii suggest a resting state re-activated by ferredoxin.
Biochim Biophys Acta Bioenerg, 1864, 2023
|
|
7WZN
 
 | | PSI-LHCI from Chlamydomonas reinhardtii with bound ferredoxin | | Descriptor: | CHLOROPHYLL A, CHLOROPHYLL A ISOMER, CHLOROPHYLL B, ... | | Authors: | Kurisu, G, Gerle, C, Mitsuoka, K, Kawamoto, A, Tanaka, H. | | Deposit date: | 2022-02-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Three structures of PSI-LHCI from Chlamydomonas reinhardtii suggest a resting state re-activated by ferredoxin.
Biochim Biophys Acta Bioenerg, 1864, 2023
|
|
3AIB
 
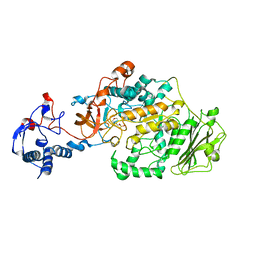 | | Crystal Structure of Glucansucrase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Glucosyltransferase-SI, ... | | Authors: | Ito, K, Ito, S, Shimamura, T, Iwata, S. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans.
J.Mol.Biol., 408, 2011
|
|
3AIC
 
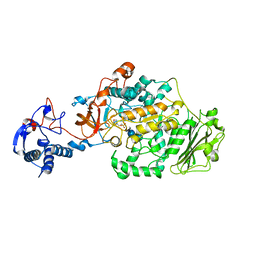 | | Crystal Structure of Glucansucrase from Streptococcus mutans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ito, K, Ito, S, Shimamura, T, Iwata, S. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans.
J.Mol.Biol., 408, 2011
|
|
3AIE
 
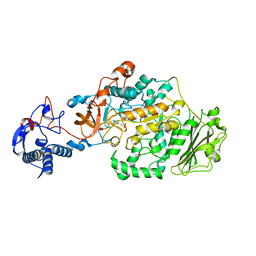 | | Crystal Structure of glucansucrase from Streptococcus mutans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Glucosyltransferase-SI | | Authors: | Ito, K, Ito, S, Shimamura, T, Iwata, S. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans.
J.Mol.Biol., 408, 2011
|
|
