6J0O
 
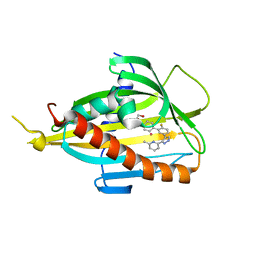 | | Crystal structure of CERT START domain in complex with compound SC1 | | Descriptor: | 2-[4-[2-fluoranyl-5-[3-(6-methylpyridin-2-yl)-1~{H}-pyrazol-4-yl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT, UNKNOWN ATOM OR ION | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-12-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
1GZQ
 
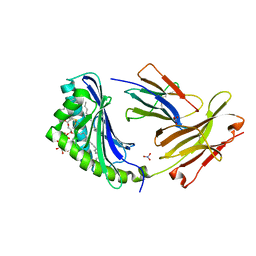 | | CD1b in complex with Phophatidylinositol | | Descriptor: | 2-[(HYDROXY{[(2R,3R,5S,6R)-2,3,4,5,6-PENTAHYDROXYCYCLOHEXYL]OXY}PHOSPHORYL)OXY]-1-[(PALMITOYLOXY)METHYL]ETHYL HEPTADECANOATE, B2-MICROGLOBULIN, DOCOSANE, ... | | Authors: | Gadola, S.D, Zaccai, N.R, Harlos, K, Shepherd, D, Ritter, G, Schmidt, R.R, Jones, E.Y, Cerundolo, V. | | Deposit date: | 2002-05-24 | | Release date: | 2002-07-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure of Human Cd1B with Bound Ligands at 2.3 A, a Maze for Alkyl Chains
Nat.Immunol., 3, 2002
|
|
1GYH
 
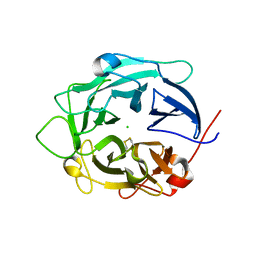 | | Structure of D158A Cellvibrio cellulosa alpha-L-arabinanase mutant | | Descriptor: | ARABINAN ENDO-1,5-ALPHA-L-ARABINOSIDASE A, CHLORIDE ION | | Authors: | Nurizzo, D, Turkenburg, J.P, Charnock, S.J, Roberts, S.M, Dodson, E.J, McKie, V.A, Taylor, E.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2002-04-23 | | Release date: | 2002-08-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Cellovibrio Cellulosa Alpha-L-Arabinanase 43A Has a Novel Five-Blade Beta-Propeller Fold
Nat.Struct.Biol., 9, 2002
|
|
5BZ6
 
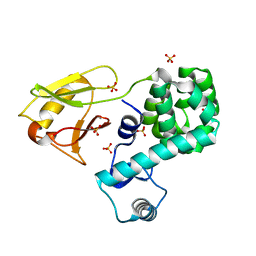 | | Crystal structure of the N-terminal domain single mutant (S92A) of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-06-11 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
6JLH
 
 | | Structure of SCGN in complex with a Snap25 peptide | | Descriptor: | CALCIUM ION, CHLORIDE ION, Secretagogin, ... | | Authors: | Qin, J, Sun, Q, Jia, D. | | Deposit date: | 2019-03-05 | | Release date: | 2020-03-11 | | Last modified: | 2020-04-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural and mechanistic insights into secretagogin-mediated exocytosis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5C1V
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CATALYTIC SUBUNIT OF HUMAN CALCINEURIN | | Descriptor: | FE (III) ION, PHOSPHATE ION, Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform, ... | | Authors: | Guasch, A, Fita, I, Perez-Luque, R, Aparicio, D, Aranguren-Ibanez, A, Perez-Riba, M. | | Deposit date: | 2015-06-15 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Calcineurin Undergoes a Conformational Switch Evoked via Peptidyl-Prolyl Isomerization.
Plos One, 10, 2015
|
|
1GSI
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH THYMIDINE MONOPHOSPHATE (TMP) | | Descriptor: | ACETATE ION, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Ursby, T, Weik, M, Fioravanti, E, Delarue, M, Goeldner, M, Bourgeois, D. | | Deposit date: | 2002-01-03 | | Release date: | 2002-03-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cryophotolysis of Caged Compounds: A Technique for Trapping Intermediate States in Protein Crystals
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1HH1
 
 | | THE STRUCTURE OF HJC, A HOLLIDAY JUNCTION RESOLVING ENZYME FROM SULFOLOBUS SOLFATARICUS | | Descriptor: | HOLLIDAY JUNCTION RESOLVING ENZYME HJC | | Authors: | Bond, C.S, Kvaratskhelia, M, Richard, D, White, M.F, Hunter, W.N. | | Deposit date: | 2000-12-18 | | Release date: | 2001-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Hjc, a Holliday Junction Resolvase, from Sulfolobus Solfataricus
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
6IZV
 
 | | Averaged strand structure of a 15-stranded ParM filament from Clostridium botulinum | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative plasmid segregation protein ParM | | Authors: | Koh, F, Narita, A, Lee, L.J, Tan, Y.Z, Dandey, V.P, Tanaka, K, Popp, D, Robinson, R.C. | | Deposit date: | 2018-12-20 | | Release date: | 2019-06-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structure of a 15-stranded actin-like filament from Clostridium botulinum.
Nat Commun, 10, 2019
|
|
5C83
 
 | | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Compound 21 | | Descriptor: | (2R,5R)-4-[2-(6-benzyl-3,3-dimethyl-2,3-dihydro-1H-pyrrolo[3,2-c]pyridin-1-yl)-2-oxoethyl]-5-(methoxymethyl)-2-methylpiperazin-1-ium, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Chessari, G, Buck, I.M, Day, J.E.H, Day, P.J, Iqbal, A, Johnson, C.N, Lewis, E.J, Martins, V, Miller, D, Reader, M, Rees, D.C, Rich, S.J, Tamanini, E, Vitorino, M, Ward, G.A, Williams, P.A, Williams, G, Wilsher, N.E, Woolford, A.J.-A. | | Deposit date: | 2015-06-25 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Discovery of a Non-Alanine Lead Series with Dual Activity Against cIAP1 and XIAP.
J.Med.Chem., 58, 2015
|
|
6YEV
 
 | |
5C5Z
 
 | | Crystal structure analysis of c4763, a uropathogenic E. coli-specific protein | | Descriptor: | Glutamyl-tRNA amidotransferase | | Authors: | Kim, H, Choi, J, Kim, D, Kim, K.K. | | Deposit date: | 2015-06-22 | | Release date: | 2015-08-19 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure analysis of c4763, a uropathogenic Escherichia coli-specific protein.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5C66
 
 | |
1H8B
 
 | | EF-hands 3,4 from alpha-actinin / Z-repeat 7 from titin | | Descriptor: | ALPHA-ACTININ 2, SKELETAL MUSCLE ISOFORM, TITIN | | Authors: | Atkinson, R.A, Joseph, C, Kelly, G, Muskett, F.W, Frenkiel, T.A, Nietlispach, D, Pastore, A. | | Deposit date: | 2001-02-01 | | Release date: | 2001-08-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ca2+-Independent Binding of an EF-Hand Domain to a Novel Motif in the Alpha-Actinin-Titin Complex
Nat.Struct.Biol., 8, 2001
|
|
6YM5
 
 | | Crystal structure of BAY-091 with PIP4K2A | | Descriptor: | (2~{R})-2-[[3-cyano-2-[4-(2-fluoranyl-3-methyl-phenyl)phenyl]-1,7-naphthyridin-4-yl]amino]butanoic acid, PHOSPHATE ION, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | Authors: | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
6YHT
 
 | | A lid blocking mechanism of a cone snail toxin revealed at the atomic level | | Descriptor: | CITRIC ACID, Conk-C1, SULFATE ION | | Authors: | Saikia, C, Altman-Gueta, H, Dym, O, Frolow, F, Gurevitz, M, Gordon, D, Reuveny, E, Karbat, I. | | Deposit date: | 2020-03-31 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Molecular Lid Mechanism of K + Channel Blocker Action Revealed by a Cone Peptide.
J.Mol.Biol., 433, 2021
|
|
6JF1
 
 | | Crystal structure of the substrate binding protein of a methionine transporter from Streptococcus pneumoniae | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jha, B, Vyas, R, Bhushan, J, Sehgal, D, Biswal, B.K. | | Deposit date: | 2019-02-07 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the substrate specificity of SP_0149, the substrate-binding protein of a methionine ABC transporter from Streptococcus pneumoniae.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
1I16
 
 | | STRUCTURE OF INTERLEUKIN 16: IMPLICATIONS FOR FUNCTION, NMR, 20 STRUCTURES | | Descriptor: | INTERLEUKIN 16 | | Authors: | Muehlhahn, P, Zweckstetter, M, Georgescu, J, Ciosto, C, Renner, C, Lanzendoerfer, M, Lang, K, Ambrosius, D, Baier, M, Kurth, R, Holak, T.A. | | Deposit date: | 1998-05-20 | | Release date: | 1999-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of interleukin 16 resembles a PDZ domain with an occluded peptide binding site.
Nat.Struct.Biol., 5, 1998
|
|
6YHY
 
 | | A lid blocking mechanism of a cone snail toxin revealed at the atomic level | | Descriptor: | Conk-S1 | | Authors: | Saikia, C, Altman-Gueta, H, Dym, O, Frolow, F, Gurevitz, M, Gordon, D, Reuveny, E, Karbat, I. | | Deposit date: | 2020-03-31 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Molecular Lid Mechanism of K + Channel Blocker Action Revealed by a Cone Peptide.
J.Mol.Biol., 433, 2021
|
|
5CFS
 
 | | Crystal Structure of ANT(2")-Ia in complex with AMPCPP and tobramycin | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, AAD(2''),Gentamicin 2''-nucleotidyltransferase,Gentamicin resistance protein, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Rodionov, D, Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2015-07-08 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis of the Tobramycin and Gentamicin Clinical Resistome Reveals Limitations for Next-generation Aminoglycoside Design.
Acs Chem.Biol., 11, 2016
|
|
6JHR
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F6 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-18 | | Release date: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
1I4K
 
 | | CRYSTAL STRUCTURE OF AN SM-LIKE PROTEIN (AF-SM1) FROM ARCHAEOGLOBUS FULGIDUS AT 2.5A RESOLUTION | | Descriptor: | CITRIC ACID, PUTATIVE SNRNP SM-LIKE PROTEIN | | Authors: | Toro, I, Thore, S, Mayer, C, Basquin, J, Seraphin, B, Suck, D. | | Deposit date: | 2001-02-22 | | Release date: | 2001-08-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex.
EMBO J., 20, 2001
|
|
6IZR
 
 | | Whole structure of a 15-stranded ParM filament from Clostridium botulinum | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative plasmid segregation protein ParM | | Authors: | Koh, F, Narita, A, Lee, L.J, Tan, Y.Z, Dandey, V.P, Tanaka, K, Popp, D, Robinson, R.C. | | Deposit date: | 2018-12-20 | | Release date: | 2019-06-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | The structure of a 15-stranded actin-like filament from Clostridium botulinum.
Nat Commun, 10, 2019
|
|
6JMF
 
 | |
1I5H
 
 | |
