1Q7O
 
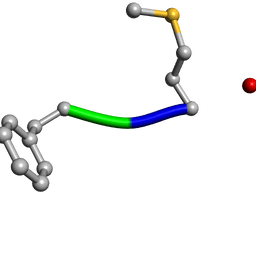 | | Determination of f-MLF-OH Peptide Structure with solid-state magic-angle spinning NMR Spectroscopy | | Descriptor: | chemotactic peptide | | Authors: | Rienstra, C.M, Tucker-Kellogg, L, Jaroniec, C.P, Hohwy, M, Reif, B, McMahon, M.T, Tidor, B, Lozano-Perez, T, Griffin, R.G. | | Deposit date: | 2003-08-19 | | Release date: | 2003-09-09 | | Last modified: | 2022-03-02 | | Method: | SOLID-STATE NMR | | Cite: | De novo determination of peptide structure with solid-state magic-angle spinning NMR Spectroscopy
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KDR
 
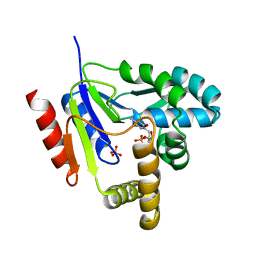 | | CYTIDINE MONOPHOSPHATE KINASE FROM E.COLI IN COMPLEX WITH ARA-CYTIDINE MONOPHOSPHATE | | Descriptor: | CYTIDYLATE KINASE, CYTOSINE ARABINOSE-5'-PHOSPHATE, SULFATE ION | | Authors: | Bertrand, T, Briozzo, P, Assairi, L, Ofiteru, A, Bucurenci, N, Munier-Lehmann, H, Golinelli-Pimpaneau, B, Barzu, O, Gilles, A.M. | | Deposit date: | 2001-11-13 | | Release date: | 2002-01-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Sugar specificity of bacterial CMP kinases as revealed by crystal structures and mutagenesis of Escherichia coli enzyme.
J.Mol.Biol., 315, 2002
|
|
1KU1
 
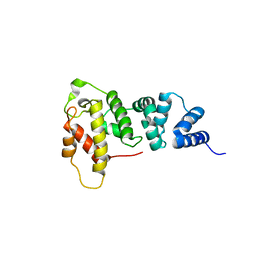 | | Crystal Structure of the Sec7 Domain of Yeast GEA2 | | Descriptor: | ARF guanine-nucleotide exchange factor 2 | | Authors: | Renault, L, Christova, P, Guibert, B, Pasqualato, S, Cherfils, J. | | Deposit date: | 2002-01-20 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mechanism of domain closure of Sec7 domains and role in BFA sensitivity.
Biochemistry, 41, 2002
|
|
1QC8
 
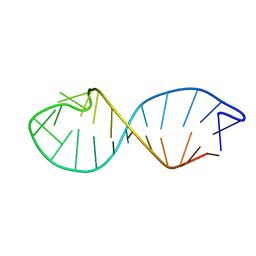 | | NMR STRUCTURE OF TAU EXON 10 SPLICING REGULATORY ELEMENT RNA | | Descriptor: | TAU EXON 10 SPLICING REGULATORY ELEMENT RNA | | Authors: | Varani, L, Spillantini, M.G, Klug, A, Goedert, M, Varani, G. | | Deposit date: | 1999-05-18 | | Release date: | 1999-08-31 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of tau exon 10 splicing regulatory element RNA and destabilization by mutations of frontotemporal dementia and parkinsonism linked to chromosome 17.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1IMU
 
 | | Solution Structure of HI0257, a Ribosome Binding Protein | | Descriptor: | HYPOTHETICAL PROTEIN HI0257 | | Authors: | Parsons, L, Eisenstein, E, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-11 | | Release date: | 2001-10-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of HI0257, a bacterial ribosome binding protein.
Biochemistry, 40, 2001
|
|
1Q6Y
 
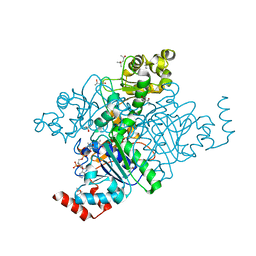 | | Hypothetical protein YfdW from E. coli bound to Coenzyme A | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COENZYME A, Hypothetical protein yfdW | | Authors: | Gogos, A, Gorman, J, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-08-14 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of Escherichia coli YfdW, a type III CoA transferase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1QQT
 
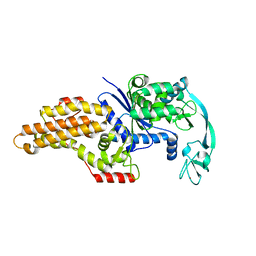 | | METHIONYL-TRNA SYNTHETASE FROM ESCHERICHIA COLI | | Descriptor: | METHIONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Mechulam, Y, Schmitt, E, Maveyraud, L, Zelwer, C, Nureki, O, Yokoyama, S, Konno, M, Blanquet, S. | | Deposit date: | 1999-06-08 | | Release date: | 2000-01-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Escherichia coli methionyl-tRNA synthetase highlights species-specific features.
J.Mol.Biol., 294, 1999
|
|
5VEW
 
 | | Structure of the human GLP-1 receptor complex with PF-06372222 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Glucagon-like peptide 1 receptor,Endolysin chimera, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine, ... | | Authors: | Song, G, Yang, D, Wang, Y, Graaf, C.D, Zhou, Q, Jiang, S, Liu, K, Cai, X, Dai, A, Lin, G, Liu, D, Wu, F, Wu, Y, Zhao, S, Ye, L, Han, G.W, Lau, J, Wu, B, Hanson, M.A, Liu, Z.-J, Wang, M.-W, Stevens, R.C. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators.
Nature, 546, 2017
|
|
1J39
 
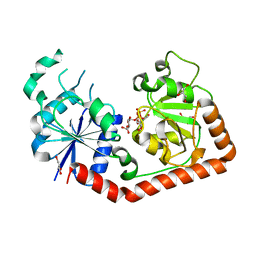 | | Crystal Structure of T4 phage BGT in complex with its UDP-glucose substrate | | Descriptor: | DNA beta-glucosyltransferase, GLYCEROL, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Lariviere, L, Morera, S. | | Deposit date: | 2003-01-21 | | Release date: | 2003-08-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of the T4 phage beta-glucosyltransferase and the D100A mutant in complex with UDP-glucose: glucose binding and identification of the catalytic base for a direct displacement mechanism.
J.Mol.Biol., 330, 2003
|
|
5VX9
 
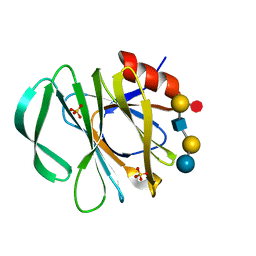 | | VP8* of P[6] Human Rotavirus RV3 in complex with LNFP1 | | Descriptor: | Outer capsid protein VP4, SULFATE ION, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hu, L, Venkataram Prasad, B.V. | | Deposit date: | 2017-05-23 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Glycan recognition in globally dominant human rotaviruses.
Nat Commun, 9, 2018
|
|
5VJX
 
 | | Crystal structure of the CLOCK Transcription Domain Exon19 in Complex with a Repressor | | Descriptor: | CLOCK-interacting pacemaker, Circadian locomoter output cycles protein kaput | | Authors: | Hou, Z, Su, L, Pei, J, Grishin, N.V, Zhang, H. | | Deposit date: | 2017-04-20 | | Release date: | 2017-12-06 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Crystal Structure of the CLOCK Transactivation Domain Exon19 in Complex with a Repressor.
Structure, 25, 2017
|
|
1QVI
 
 | | Crystal structure of scallop myosin S1 in the pre-power stroke state to 2.6 Angstrom resolution: flexibility and function in the head | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Gourinath, S, Himmel, D.M, Brown, J.H, Reshetnikova, L, Szent-Gyrgyi, A.G, Cohen, C. | | Deposit date: | 2003-08-27 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of scallop Myosin s1 in the pre-power stroke state to 2.6 a resolution: flexibility and function in the head.
Structure, 11, 2003
|
|
5VIP
 
 | |
1Q9O
 
 | | S45-18 Fab Unliganded | | Descriptor: | MAGNESIUM ION, S45-2 Fab (IgG1k) heavy chain, S45-2 Fab (IgG1k) light chain | | Authors: | Nguyen, H.P, Seto, N.O, MacKenzie, C.R, Brade, L, Kosma, P, Brade, H, Evans, S.V. | | Deposit date: | 2003-08-25 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Germline antibody recognition of distinct carbohydrate epitopes.
Nat.Struct.Biol., 10, 2003
|
|
5W0M
 
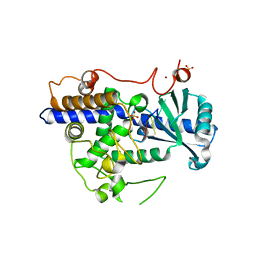 | | Structure of human TUT7 catalytic module (CM) in complex with U5 RNA | | Descriptor: | IODIDE ION, SULFATE ION, Terminal uridylyltransferase 7, ... | | Authors: | Faehnle, C.R, Walleshauser, J, Joshua-Tor, L. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Multi-domain utilization by TUT4 and TUT7 in control of let-7 biogenesis.
Nat. Struct. Mol. Biol., 24, 2017
|
|
1QXX
 
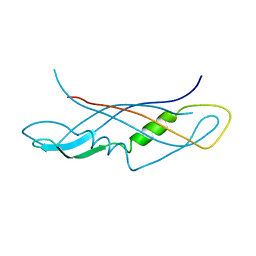 | | CRYSTAL STRUCTURE OF THE C-TERMINAL DOMAIN OF TONB | | Descriptor: | TonB protein | | Authors: | Koedding, J, Howard, P, Kaufmann, L, Polzer, P, Lustig, A, Welte, W. | | Deposit date: | 2003-09-09 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dimerization of TonB is not essential for its binding to the outer membrane siderophore receptor FhuA of Escherichia coli.
J.Biol.Chem., 279, 2004
|
|
1J4H
 
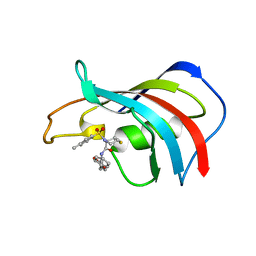 | | crystal structure analysis of the FKBP12 complexed with 000107 small molecule | | Descriptor: | 3-PHENYL-2-{[4-(TOLUENE-4-SULFONYL)-THIOMORPHOLINE-3-CARBONYL]-AMINO}-PROPIONIC ACID ETHYL ESTER, FKBP12 | | Authors: | Li, P, Ding, Y, Wang, L, Wu, B, Shu, C, Li, S, Shen, B, Rao, Z. | | Deposit date: | 2001-09-30 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and structure-based study of new potential FKBP12 inhibitors.
Biophys.J., 85, 2003
|
|
4DXZ
 
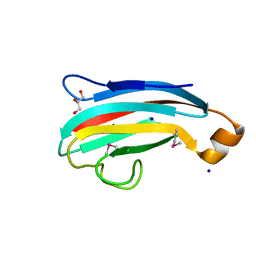 | | crystal structure of a PliG-Ec mutant, a periplasmic lysozyme inhibitor of g-type lysozyme from Escherichia coli | | Descriptor: | GLYCEROL, Inhibitor of g-type lysozyme, SODIUM ION | | Authors: | Leysen, S, Vanheuverzwijn, S, Van Asten, K, Vanderkelen, L, Michiels, C.W, Strelkov, S.V. | | Deposit date: | 2012-02-28 | | Release date: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | crystal structure of a PliG-Ec mutant, a periplasmic lysozyme inhibitor of g-type lysozyme from Escherichia coli
To be Published
|
|
1Q8I
 
 | | Crystal structure of ESCHERICHIA coli DNA Polymerase II | | Descriptor: | DNA polymerase II | | Authors: | Brunzelle, J.S, Muchmore, C.R.A, Mashhoon, N, Blair-Johnson, M, Shuvalova, L, Goodman, M.F, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-08-21 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Escherichia Coli DNA Polymerase II
To be Published
|
|
1IDI
 
 | | THE NMR SOLUTION STRUCTURE OF ALPHA-BUNGAROTOXIN | | Descriptor: | ALPHA-BUNGAROTOXIN | | Authors: | Zeng, H, Moise, L, Grant, M.A, Hawrot, E. | | Deposit date: | 2001-04-04 | | Release date: | 2001-04-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the complex formed between alpha-bungarotoxin and an 18-mer cognate peptide derived from the alpha 1 subunit of the nicotinic acetylcholine receptor from Torpedo californica.
J.Biol.Chem., 276, 2001
|
|
1QKK
 
 | | Crystal structure of the receiver domain and linker region of DctD from Sinorhizobium meliloti | | Descriptor: | C4-DICARBOXYLATE TRANSPORT TRANSCRIPTIONAL REGULATORY PROTEIN | | Authors: | Meyer, M.G, Park, S, Zeringue, L, Staley, M, Mckinstry, M, Kaufman, R.I, Zhang, H, Yan, D, Yennawar, N, Farber, G.K, Nixon, B.T. | | Deposit date: | 1999-07-23 | | Release date: | 2000-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A dimeric two-component receiver domain inhibits the sigma54-dependent ATPase in DctD.
Faseb J., 15, 2001
|
|
5VIT
 
 | |
1QNC
 
 | | Crystal structure of the A(-31) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | Descriptor: | DNA (5'-D(*GP*CP*AP*AP*TP*AP*AP*AP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*TP*TP*TP*AP*TP*TP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | Authors: | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | Deposit date: | 1999-10-14 | | Release date: | 2000-02-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
1IGN
 
 | | DNA-BINDING DOMAIN OF RAP1 IN COMPLEX WITH TELOMERIC DNA SITE | | Descriptor: | DNA (5'-D(*CP*CP*GP*CP*AP*CP*AP*CP*CP*CP*AP*CP*AP*CP*AP*CP*C P*AP*G)-3'), DNA (5'-D(*CP*CP*TP*GP*GP*TP*GP*TP*GP*TP*GP*GP*GP*TP*GP*TP*G P*CP*G)-3'), PROTEIN (RAP1) | | Authors: | Koenig, P, Giraldo, R, Chapman, L, Rhodes, D. | | Deposit date: | 1996-02-29 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA.
Cell(Cambridge,Mass.), 85, 1996
|
|
1QTK
 
 | | CRYSTAL STRUCTURE OF HEW LYSOZYME UNDER PRESSURE OF KRYPTON (55 BAR) | | Descriptor: | CHLORIDE ION, KRYPTON, LYSOZYME, ... | | Authors: | Prange, T, Schiltz, M, Pernot, L, Colloc'h, N, Longhi, S, Bourguet, W, Fourme, R. | | Deposit date: | 1999-06-28 | | Release date: | 1999-07-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Exploring hydrophobic sites in proteins with xenon or krypton.
Proteins, 30, 1998
|
|
