6NJN
 
 | |
2IO8
 
 | | E. coli Bifunctional glutathionylspermidine synthetase/amidase Incomplex with Mg2+ and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional glutathionylspermidine synthetase/amidase, CYSTEINE, ... | | Authors: | Pai, C.H, Chiang, B.Y, Ko, T.P, Chou, C.C, Chong, C.M, Yen, F.J, Coward, J.K, Wang, A.H.-J, Lin, C.H. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dual binding sites for translocation catalysis by Escherichia coli glutathionylspermidine synthetase
Embo J., 25, 2006
|
|
2IOA
 
 | | E. coli Bifunctional glutathionylspermidine synthetase/amidase Incomplex with Mg2+ and ADP and phosphinate inhibitor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional glutathionylspermidine synthetase/amidase, D-GAMMA-GLUTAMYL-N-{[(R)-{4-[(4-AMINOBUTYL)AMINO]BUTYL}(PHOSPHONOOXY)PHOSPHORYL]METHYL}-D-ALANINAMIDE, ... | | Authors: | Pai, C.H, Chiang, B.Y, Ko, T.P, Chou, C.C, Chong, C.M, Yen, F.J, Coward, J.K, Wang, A.H.-J, Lin, C.H. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual binding sites for translocation catalysis by Escherichia coli glutathionylspermidine synthetase
Embo J., 25, 2006
|
|
2IO9
 
 | | E. coli Bifunctional glutathionylspermidine synthetase/amidase Incomplex with Mg2+ ,GSH and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional glutathionylspermidine synthetase/amidase, GLUTATHIONE, ... | | Authors: | Pai, C.H, Chiang, B.Y, Ko, T.P, Chou, C.C, Chong, C.M, Yen, F.J, Coward, J.K, Wang, A.H.-J, Lin, C.H. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dual binding sites for translocation catalysis by Escherichia coli glutathionylspermidine synthetase
Embo J., 25, 2006
|
|
2IOB
 
 | | E. coli Bifunctional glutathionylspermidine synthetase/amidase Apo protein | | Descriptor: | Bifunctional glutathionylspermidine synthetase/amidase | | Authors: | Pai, C.H, Chiang, B.Y, Ko, T.P, Chou, C.C, Chong, C.M, Yen, F.J, Coward, J.K, Wang, A.H.-J, Lin, C.H. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dual binding sites for translocation catalysis by Escherichia coli glutathionylspermidine synthetase
Embo J., 25, 2006
|
|
2IIZ
 
 | |
2IO7
 
 | | E. coli Bifunctional glutathionylspermidine synthetase/amidase Incomplex with Mg2+ and AMPPNP | | Descriptor: | Bifunctional glutathionylspermidine synthetase/amidase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Pai, C.H, Chiang, B.Y, Ko, T.P, Chong, C.M, Yen, F.J, Coward, J.K, Wang, A.H.-J, Lin, C.H. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dual binding sites for translocation catalysis by Escherichia coli glutathionylspermidine synthetase
Embo J., 25, 2006
|
|
7E24
 
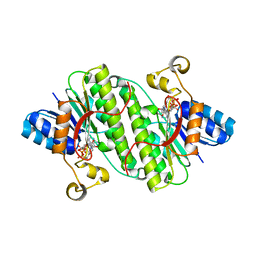 | |
7E28
 
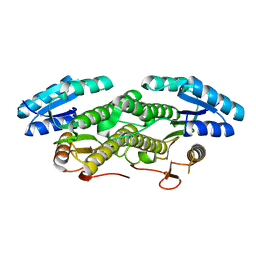 | |
6INB
 
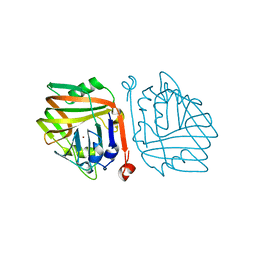 | |
6INC
 
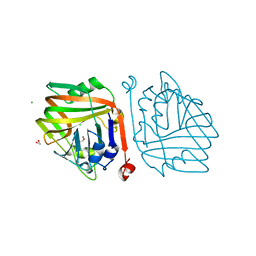 | | Crystal structure of an acetolactate decarboxylase from Klebsiella pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, Alpha-acetolactate decarboxylase, CHLORIDE ION, ... | | Authors: | Wu, W, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Structural characterization of an acetolactate decarboxylase from Klebsiella pneumoniae
Biochem. Biophys. Res. Commun., 509, 2019
|
|
3C0F
 
 | | Crystal Structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source | | Descriptor: | Uncharacterized protein AF_1514 | | Authors: | Li, Y, Bahti, P, Shaw, N, Song, G, Yin, J, Zhu, J.-Y, Zhang, H, Xu, H, Wang, B.-C, Liu, Z.-J. | | Deposit date: | 2008-01-20 | | Release date: | 2008-02-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source.
Proteins, 71, 2008
|
|
5LUI
 
 | | Structure of cutinase 1 from Thermobifida cellulosilytica | | Descriptor: | CHLORIDE ION, Cutinase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hromic, A, Lyskowski, A, Gruber, K. | | Deposit date: | 2016-09-08 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Small cause, large effect: Structural characterization of cutinases from Thermobifida cellulosilytica.
Biotechnol. Bioeng., 114, 2017
|
|
5LUK
 
 | | Structure of a double variant of cutinase 2 from Thermobifida cellulosilytica | | Descriptor: | CHLORIDE ION, Cutinase 2, MAGNESIUM ION | | Authors: | Hromic, A, Lyskowski, A, Gruber, K. | | Deposit date: | 2016-09-09 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Small cause, large effect: Structural characterization of cutinases from Thermobifida cellulosilytica.
Biotechnol. Bioeng., 114, 2017
|
|
5LUJ
 
 | |
5YKR
 
 | |
5YKT
 
 | |
6LLN
 
 | |
4WG5
 
 | |
4WG3
 
 | |
6DCV
 
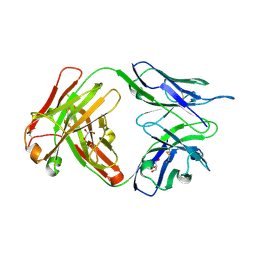 | | Crystal structure of human anti-tau antibody CBTAU-27.1 | | Descriptor: | GLYCEROL, Light chain of CBTAU27.1 Fab, heavy chain of CBTAU-27.1 Fab | | Authors: | Zhu, X, Zhang, H, Wilson, I.A. | | Deposit date: | 2018-05-08 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A common antigenic motif recognized by naturally occurring human VH5-51/VL4-1 anti-tau antibodies with distinct functionalities.
Acta Neuropathol Commun, 6, 2018
|
|
4WG4
 
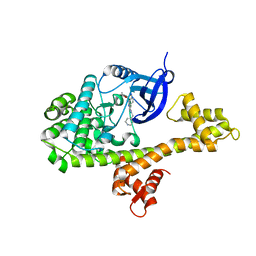 | |
4WSJ
 
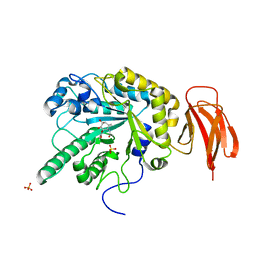 | | Crystal structure of a bacterial fucodiase in complex with 1-((1R,2R,3R,4R,5R,6R)-2,3,4-trihydroxy-5-methyl-7-azabicyclo[4.1.0]heptan-7-yl)ethan-1-one | | Descriptor: | Alpha-L-fucosidase, N-[(1S,2R,3R,4S,5R)-3,4,5-trihydroxy-2-methylcyclohexyl]acetamide, SULFATE ION | | Authors: | Davies, G.J, Wright, D.W. | | Deposit date: | 2014-10-28 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | In vitroandin vivocomparative and competitive activity-based protein profiling of GH29 alpha-l-fucosidases.
Chem Sci, 6, 2015
|
|
4WSK
 
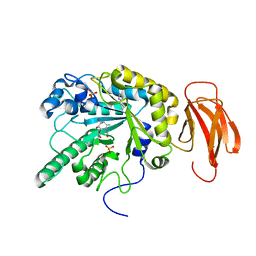 | | Crystal structure of a bacterial fucosidase with phenyl((1R,2R,3R,4R,5R,6R)-2,3,4-trihydroxy-5-methyl-7-azabicyclo[4.1.0]heptan-7-yl)methanone | | Descriptor: | Alpha-L-fucosidase, IMIDAZOLE, N-[(1S,2R,3R,4S,5R)-3,4,5-trihydroxy-2-methylcyclohexyl]benzamide, ... | | Authors: | Davies, G.J. | | Deposit date: | 2014-10-28 | | Release date: | 2014-11-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | In vitroandin vivocomparative and competitive activity-based protein profiling of GH29 alpha-l-fucosidases.
Chem Sci, 6, 2015
|
|
3NG7
 
 | |
