3GUV
 
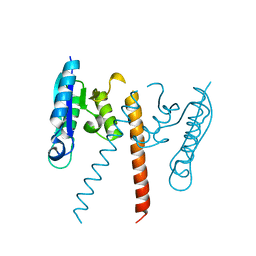 | | Crystal structure of a resolvase family site-specific recombinase from Streptococcus pneumoniae | | Descriptor: | Site-specific recombinase, resolvase family protein | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Do, J, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-30 | | Release date: | 2009-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a resolvase family site-specific recombinase from Streptococcus pneumoniae
To be Published
|
|
1P8L
 
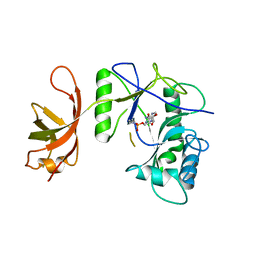 | | New Crystal Structure of Chlorella Virus DNA Ligase-Adenylate | | Descriptor: | ADENOSINE MONOPHOSPHATE, PBCV-1 DNA ligase | | Authors: | Odell, M, Malinina, L, Teplova, M, Shuman, S. | | Deposit date: | 2003-05-07 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Analysis of the DNA Joining Repertoire of Chlorella Virus DNA ligase and a New Crystal Structure of the Ligase-Adenylate Intermediate
Nucleic Acids Res., 31, 2003
|
|
3CTA
 
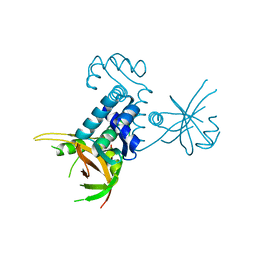 | | Crystal structure of riboflavin kinase from Thermoplasma acidophilum | | Descriptor: | Riboflavin kinase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Mendoza, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-11 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of riboflavin kinase from Thermoplasma acidophilum.
To be Published
|
|
1OS9
 
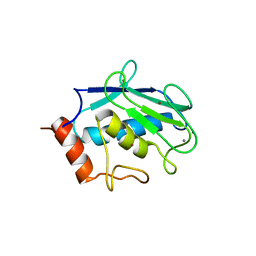 | | Binary enzyme-product complexes of human MMP12 | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, ZINC ION | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Mangani, S, Terni, B. | | Deposit date: | 2003-03-19 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray Structures of Binary and Ternary Enzyme-Product-Inhibitor Complexes of Matrix Metalloproteinases
Angew.Chem.Int.Ed.Engl., 42, 2003
|
|
1Q8O
 
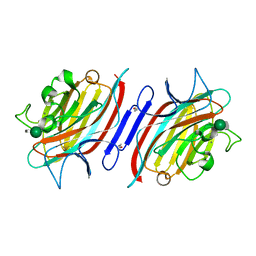 | | Pterocartpus angolensis lectin PAL in complex with the dimmanoside Man(alpha1-2)Man | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, alpha-D-mannopyranose-(1-2)-methyl alpha-D-mannopyranoside, ... | | Authors: | Loris, R, Van Walle, I, De Greve, H, Beeckmans, S, Deboeck, F, Wyns, L, Bouckaert, J. | | Deposit date: | 2003-08-22 | | Release date: | 2004-02-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Oligomannose Recognition by the Pterocarpus angolensis Seed Lectin
J.Mol.Biol., 335, 2004
|
|
1Q8S
 
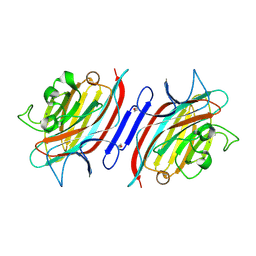 | | Pterocarpus angolensis lectin (PAL) in complex with the dimannoside Man(alpha1-6)Man | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, alpha-D-mannopyranose-(1-6)-methyl alpha-D-mannopyranoside, ... | | Authors: | Loris, R, Van Walle, I, De Greve, H, Beeckmans, S, DeBoeck, F, Wyns, L, Bouckaert, J. | | Deposit date: | 2003-08-22 | | Release date: | 2004-02-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis of Oligomannose Recognition by the Pterocarpus angolensis Seed Lectin
J.Mol.Biol., 335, 2004
|
|
1QSK
 
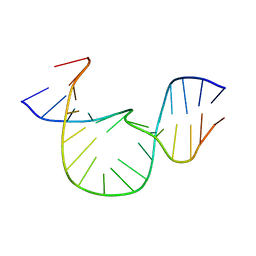 | | NMR-DERIVED SOLUTION STRUCTURE OF A FIVE-ADENINE BULGE LOOP WITHIN A DNA DUPLEX | | Descriptor: | 5'-D(CP*GP*TP*AP*GP*CP*CP*GP*AP*TP*GP*CP)-3', 5'-D(GP*CP*AP*TP*CP*GP*AP*AP*AP*AP*AP*GP*CP*TP*AP*CP*GP)-3' | | Authors: | Dornberger, U, Hillisch, A, Gollmick, F, Fritzsche, H, Diekmann, S. | | Deposit date: | 1999-06-22 | | Release date: | 1999-11-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a five-adenine bulge loop within a DNA duplex.
Biochemistry, 38, 1999
|
|
6XZF
 
 | |
6XYF
 
 | | Nanobody 22 | | Descriptor: | Nanobody 22, SODIUM ION | | Authors: | Pompidor, G, Zimmermann, S, Loew, C, Schneider, T. | | Deposit date: | 2020-01-30 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.11097 Å) | | Cite: | Engineered nanobodies with a lanthanide binding motif for crystallographic phasing
To Be Published
|
|
1R4D
 
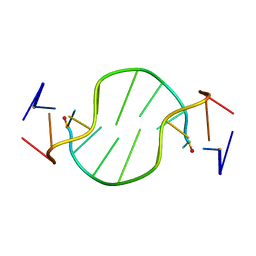 | | Solution structure of the chimeric L/D DNA oligonucleotide d(C8metGCGC(L)G(L)CGCG)2 | | Descriptor: | 5'-D(*CP*(8MG)P*CP*GP*(0DC)P*(0DG)P*CP*GP*CP*G)-3' | | Authors: | Cherrak, I, Mauffret, O, Santamaria, F, Rayner, B, Hocquet, A, Ghomi, M, Fermandjian, S. | | Deposit date: | 2003-10-06 | | Release date: | 2003-10-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | L-nucleotides and 8-methylguanine of d(C1m8G2C3G4C5LG6LC7G8C9G10)2 act cooperatively to promote a left-handed helix under physiological salt conditions.
Nucleic Acids Res., 31, 2003
|
|
6XYM
 
 | | Nbe-LBM | | Descriptor: | Nbe-LBM, TERBIUM(III) ION | | Authors: | Pompidor, G, Zimmermann, S, Loew, C, Schneider, T. | | Deposit date: | 2020-01-30 | | Release date: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Engineered nanobodies with a lanthanide binding motif for crystallographic phasing
To Be Published
|
|
1RJC
 
 | | Crystal structure of the camelid single domain antibody cAb-Lys2 in complex with hen egg white lysozyme | | Descriptor: | GLYCEROL, Lysozyme C, PHOSPHATE ION, ... | | Authors: | De Genst, E, Silence, K, Ghahroudi, M.A, Decanniere, K, Loris, R, Kinne, J, Wyns, L, Muyldermans, S. | | Deposit date: | 2003-11-19 | | Release date: | 2005-02-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Strong in vivo maturation compensates for structurally restricted H3 loops in antibody repertoires.
J.Biol.Chem., 280, 2005
|
|
1QX2
 
 | | X-ray Structure of Calcium-loaded Calbindomodulin (A Calbindin D9k Re-engineered to Undergo a Conformational Opening) at 1.44 A Resolution | | Descriptor: | CALCIUM ION, Vitamin D-dependent calcium-binding protein, intestinal, ... | | Authors: | Bunick, C.G, Nelson, M.R, Mangahas, S, Mizoue, L.S, Bunick, G.J, Chazin, W.J. | | Deposit date: | 2003-09-04 | | Release date: | 2004-05-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Designing Sequence to Control Protein Function in an EF-Hand Protein
J.Am.Chem.Soc., 126, 2004
|
|
7PXX
 
 | | The crystal structure of Leishmania major Pteridine Reductase 1 in complex with substrate folic acid | | Descriptor: | DI(HYDROXYETHYL)ETHER, FOLIC ACID, GLYCEROL, ... | | Authors: | Di Pisa, F, Dello Iacono, L, Mangani, S. | | Deposit date: | 2021-10-08 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of the ternary complex of Leishmania major pteridine reductase 1 with the cofactor NADP + /NADPH and the substrate folic acid.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
1OS2
 
 | | Ternary enzyme-product-inhibitor complexes of human MMP12 | | Descriptor: | ACETATE ION, ACETOHYDROXAMIC ACID, AZIDE ION, ... | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Mangani, S, Terni, B. | | Deposit date: | 2003-03-18 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray Structures of Binary and Ternary Enzyme-Product-Inhibitor Complexes of Matrix Metalloproteinases
Angew.Chem.Int.Ed.Engl., 42, 2003
|
|
3FV9
 
 | | Crystal structure of putative mandelate racemase/muconatelactonizing enzyme from ROSEOVARIUS NUBINHIBENS ISM complexed with magnesium | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Malashkevich, V.N, Rutter, M, Bain, K.T, Lau, C, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-15 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of putative mandelate racemase/muconatelactonizing enzyme from ROSEOVARIUS NUBINHIBENS ISM complexed with magnesium
to be published
|
|
3FVD
 
 | | Crystal structure of a member of enolase superfamily from ROSEOVARIUS NUBINHIBENS ISM complexed with magnesium | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Malashkevich, V.N, Rutter, M, Bain, K.T, Lau, C, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-15 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a member of enolase superfamily from ROSEOVARIUS NUBINHIBENS ISM complexed with magnesium
to be published
|
|
3BRS
 
 | | Crystal structure of sugar transporter from Clostridium phytofermentans | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator | | Authors: | Malashkevich, V.N, Patskovsky, Y, Toro, R, Meyers, A.J, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-21 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of sugar transporter from Clostridium phytofermentans.
To be Published
|
|
6XU1
 
 | | Crystal structure of tetrameric human H215A-SAMHD1 (residues 109-626) with GTP, dAMPNPP and Mg | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Arnold, L.H, Purkiss, A.G, Kelly, G, Taylor, I.A. | | Deposit date: | 2020-01-17 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis.
Nat Commun, 11, 2020
|
|
3B40
 
 | | Crystal structure of the probable dipeptidase PvdM from Pseudomonas aeruginosa | | Descriptor: | CADMIUM ION, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Bonanno, J.B, Patskovsky, Y, Dickey, M, Bain, K.T, Mendoza, M, Fong, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-23 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the probable dipeptidase PvdM from Pseudomonas aeruginosa.
To be Published
|
|
3M4A
 
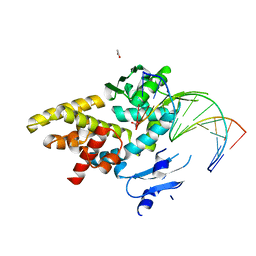 | | Crystal structure of a bacterial topoisomerase IB in complex with DNA reveals a secondary DNA binding site | | Descriptor: | ACETIC ACID, DNA (5'-D(*GP*AP*AP*TP*AP*AP*GP*GP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*CP*CP*CP*TP*TP*AP*TP*TP*C)-3'), ... | | Authors: | Patel, A, Yakovleva, L, Shuman, S, Mondragon, A. | | Deposit date: | 2010-03-10 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a bacterial topoisomerase IB in complex with DNA reveals a secondary DNA binding site.
Structure, 18, 2010
|
|
3I5R
 
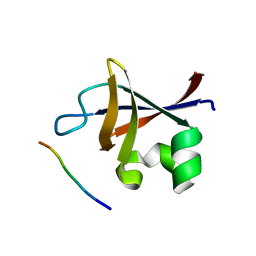 | | PI3K SH3 domain in complex with a peptide ligand | | Descriptor: | Peptide ligand, Phosphatidylinositol 3-kinase regulatory subunit alpha | | Authors: | Batra-Safferling, R, Granzin, J, Modder, S, Hoffmann, S, Willbold, D. | | Deposit date: | 2009-07-06 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of the phosphatidylinositol 3-kinase (PI3K) SH3 domain in complex with a peptide ligand: role of the anchor residue in ligand binding.
Biol.Chem., 391, 2010
|
|
3I5S
 
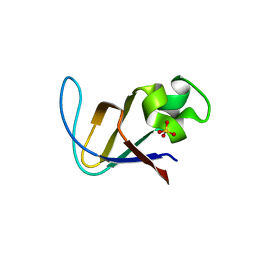 | | Crystal structure of PI3K SH3 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, SULFATE ION | | Authors: | Batra-Safferling, R, Granzin, J, Modder, S, Hoffmann, S, Willbold, D. | | Deposit date: | 2009-07-06 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies of the phosphatidylinositol 3-kinase (PI3K) SH3 domain in complex with a peptide ligand: role of the anchor residue in ligand binding.
Biol.Chem., 391, 2010
|
|
3IZJ
 
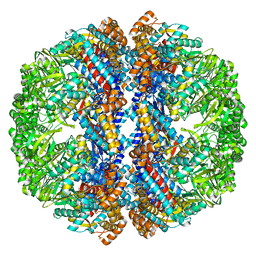 | | Mm-cpn rls with ATP and AlFx | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3BHW
 
 | | Crystal structure of an uncharacterized protein from Magnetospirillum magneticum | | Descriptor: | Uncharacterized protein | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Lau, C, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-29 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an uncharacterized protein from Magnetospirillum magneticum.
To be Published
|
|
