3ICL
 
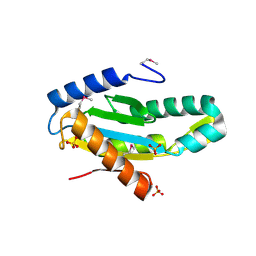 | | X-Ray Structure of Protein (EAL/GGDEF domain protein) from M.capsulatus, Northeast Structural Genomics Consortium Target McR174C | | Descriptor: | EAL/GGDEF domain protein, SULFATE ION | | Authors: | Kuzin, A, Chen, Y, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Foote, E.L, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Northeast Structural Genomics Consortium Target McR174C
To be Published
|
|
3F1T
 
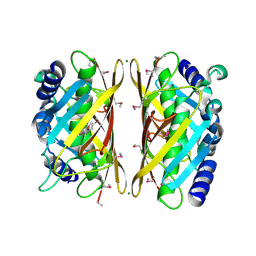 | | Crystal structure of the Q9I3C8_PSEAE protein from Pseudomonas aeruginosa. Northeast Structural Genomics Consortium target PaR319a. | | Descriptor: | MAGNESIUM ION, uncharacterized protein Q9I3C8_PSEAE | | Authors: | Vorobiev, S.M, Chen, Y, Abashidze, M, Seetharaman, J, Wang, D, Foote, E.L, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-28 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Q9I3C8_PSEAE protein from Pseudomonas aeruginosa.
To be Published
|
|
3IPF
 
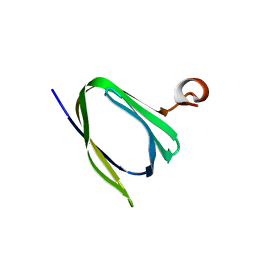 | | Crystal structure of the Q251Q8_DESHY protein from Desulfitobacterium hafniense. Northeast Structural Genomics Consortium Target DhR8c. | | Descriptor: | uncharacterized protein | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Janjua, H, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Andre, I, Rossi, P, Kennedy, M, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-08-17 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Crystal structure of the Q251Q8_DESHY protein from Desulfitobacterium hafniense.
To be Published
|
|
7F3X
 
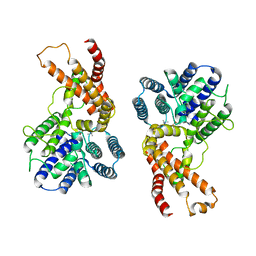 | | Lysophospholipid acyltransferase LPCAT3 in complex with lysophosphatidylcholine | | Descriptor: | LPCAT3, [2-((1-OXODODECANOXY-(2-HYDROXY-3-PROPANYL))-PHOSPHONATE-OXY)-ETHYL]-TRIMETHYLAMMONIUM | | Authors: | Zhang, Q, Yao, D, Rao, B, Li, S, Jian, L, Chen, Y, Hu, K, Xia, Y, Shen, Y, Cao, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3.
Nat Commun, 12, 2021
|
|
7EWT
 
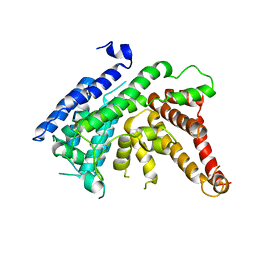 | | The crystal structure of Lysophospholipid acyltransferase LPCAT3 (MOBAT5) in its monomeric and apo form | | Descriptor: | Lysophospholipid acyltransferase 5 | | Authors: | Zhang, Q, Yao, D, Rao, B, Li, S, Jian, L, Chen, Y, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2021-05-26 | | Release date: | 2021-12-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3.
Nat Commun, 12, 2021
|
|
7F40
 
 | | Lysophospholipid acyltransferase LPCAT3 in a complex with Arachidonoyl-CoA | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, LPCAT3, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenethioate | | Authors: | Zhang, Q, Yao, D, Rao, B, Li, S, Jian, L, Chen, Y, Hu, K, Xia, Y, Shen, Y, Cao, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3.
Nat Commun, 12, 2021
|
|
5DEM
 
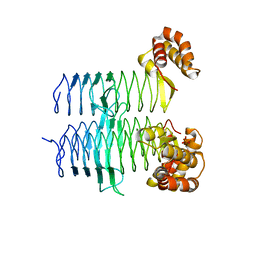 | | Structure of Pseudomonas aeruginosa LpxA | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, PHOSPHATE ION | | Authors: | Smith, E.W, Chen, Y. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structures of Pseudomonas aeruginosa LpxA Reveal the Basis for Its Substrate Selectivity.
Biochemistry, 54, 2015
|
|
5DEP
 
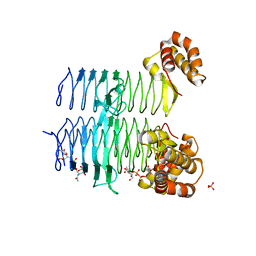 | | Structure of Pseudomonas aeruginosa LpxA in complex with UDP-GlcNAc | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, PHOSPHATE ION, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Smith, E.W, Chen, Y. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structures of Pseudomonas aeruginosa LpxA Reveal the Basis for Its Substrate Selectivity.
Biochemistry, 54, 2015
|
|
5DG3
 
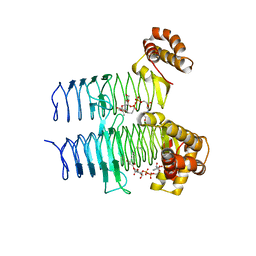 | |
5EKI
 
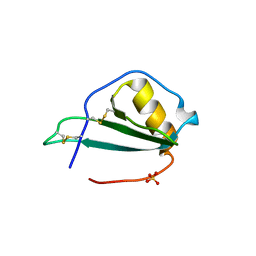 | |
7K99
 
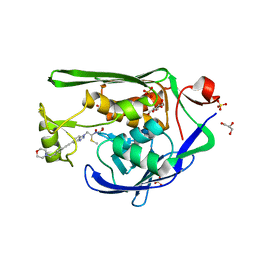 | | Crystal Structure of P. aeruginosa LpxC with N-Hydroxyformamide inhibitor 19 | | Descriptor: | (hydroxy{(1S)-1-(methylsulfanyl)-2-[5-({4-[(morpholin-4-yl)methyl]phenyl}ethynyl)-1H-benzotriazol-1-yl]ethyl}amino)methanol, GLYCEROL, SULFATE ION, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2020-09-28 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | N-Hydroxyformamide LpxC inhibitors, their in vivo efficacy in a mouse Escherichia coli infection model, and their safety in a rat hemodynamic assay.
Bioorg.Med.Chem., 28, 2020
|
|
7K9A
 
 | | Crystal Structure of P. aeruginosa LpxC with N-Hydroxyformamide inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-hydroxy-N-[(1R)-2-{5-[(4-{[2-(hydroxymethyl)-1H-imidazol-1-yl]methyl}phenyl)ethynyl]-1H-benzotriazol-1-yl}-1-(methylsulfanyl)ethyl]formamide, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2020-09-29 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-Hydroxyformamide LpxC inhibitors, their in vivo efficacy in a mouse Escherichia coli infection model, and their safety in a rat hemodynamic assay.
Bioorg.Med.Chem., 28, 2020
|
|
3KKJ
 
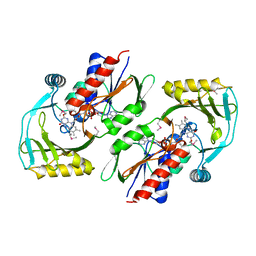 | | X-ray structure of P. syringae q888a4 Oxidoreductase at resolution 2.5A, Northeast Structural Genomics Consortium target PsR10 | | Descriptor: | Amine oxidase, flavin-containing, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kuzin, A.P, Chen, Y, Forouhar, F, Vorobiev, S, Acton, T, Ma, L.C, Xiao, R, Montelione, G.T, Hunt, J.F, Tong, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-11-05 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of P. syringae q888a4 Oxidoreductase at resolution 2.5A, Northeast Structural Genomics Consortium target PsR10
To be Published
|
|
2ES7
 
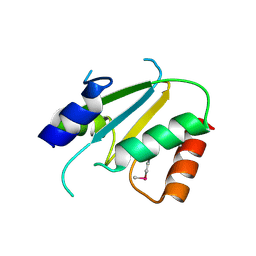 | | Crystal structure of Q8ZP25 from Salmonella typhimurium LT2. NESG TARGET STR70 | | Descriptor: | putative thiol-disulfide isomerase and thioredoxin | | Authors: | Benach, J, Su, M, Forouhar, F, Chen, Y, Ho, C.K, Janjua, H, Cunningham, K, Ma, L.-C, Rong, X, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-10-25 | | Release date: | 2005-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Q8ZP25 from Salmonella typhimurium LT2 NESG target STR70.
To be Published
|
|
2FFM
 
 | | X-Ray Crystal Structure of Protein SAV1430 from Staphylococcus aureus. Northeast Structural Genomics Consortium Target ZR18. | | Descriptor: | SAV1430 | | Authors: | Forouhar, F, Chen, Y, Jayaraman, S, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-19 | | Release date: | 2005-12-27 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of the Hypothetical Protein SAV1430 from Staphylococcus aureus, Northeast Structural Genomics ZR18.
To be Published
|
|
3KO6
 
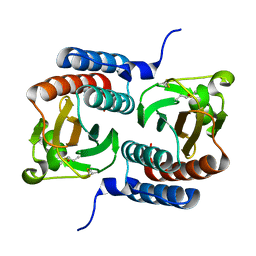 | |
3L4N
 
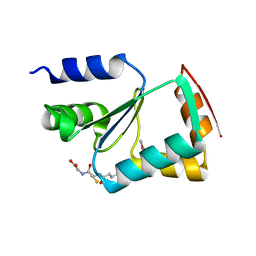 | | Crystal structure of yeast monothiol glutaredoxin Grx6 | | Descriptor: | GLUTATHIONE, Monothiol glutaredoxin-6 | | Authors: | Luo, M, Jiang, Y.-L, Ma, X.-X, He, Y.-X, Tang, Y.-J, Yu, J, Zhang, R.-G, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-04-07 | | Last modified: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and biochemical characterization of yeast monothiol glutaredoxin Grx6
J.Mol.Biol., 398, 2010
|
|
3LD7
 
 | | Crystal structure of the Lin0431 protein from Listeria innocua, Northeast Structural Genomics Consortium Target LkR112 | | Descriptor: | Lin0431 protein | | Authors: | Vorobiev, S, Chen, Y, Lee, D, Patel, D.J, Ciccosanti, C, Sahdev, S, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-12 | | Release date: | 2010-01-26 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Crystal structure of the Lin0431 protein from Listeria innocua
To be Published
|
|
3LD0
 
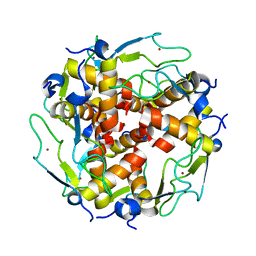 | | Crystal structure of B.licheniformis Anti-TRAP protein, an antagonist of TRAP-RNA interactions | | Descriptor: | Inhibitor of TRAP, regulated by T-BOX (Trp) sequence RtpA, MAGNESIUM ION, ... | | Authors: | Shevtsov, M.B, Chen, Y, Isupov, M.N, Gollnick, P, Antson, A.A. | | Deposit date: | 2010-01-12 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacillus licheniformis Anti-TRAP can assemble into two types of dodecameric particles with the same symmetry but inverted orientation of trimers.
J.Struct.Biol., 170, 2010
|
|
3LRQ
 
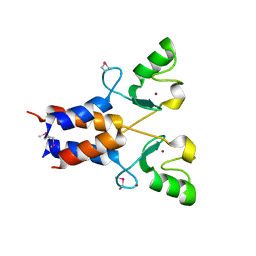 | | Crystal structure of the U-box domain of human ubiquitin-protein ligase (E3), NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET HR4604D. | | Descriptor: | E3 ubiquitin-protein ligase TRIM37, ZINC ION | | Authors: | Kuzin, A, Chen, Y, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-02-11 | | Release date: | 2010-03-23 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | E3 UBIQUITIN-PROTEIN LIGASE RING DOMAIN FROM HUMAN TRIPARTITE MOTIF-CONTAINING PROTEIN 37 (TRIM37), NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET HR4604D.
To be Published
|
|
2HZB
 
 | | X-Ray Crystal Structure of Protein BH3568 from Bacillus halodurans. Northeast Structural Genomics Consortium BhR60. | | Descriptor: | Hypothetical UPF0052 protein BH3568 | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Benach, J, Shastry, R, Conover, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-08-08 | | Release date: | 2006-08-22 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray structure of the hypothetical UPF0052 protein BH3568 from Bacillus halodurans. Northeast Structural Genomics Consortium BhR60.
To be Published
|
|
3L9E
 
 | | Crystal structures of holo and Cu-deficient Cu/ZnSOD from the silkworm Bombyx mori and the implications in Amyotrophic lateral sclerosis | | Descriptor: | Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Zhang, N.-N, He, Y.-X, Li, W.-F, Zhao, F, Yan, L.-F, Zhang, G.-Z, Teng, Y.-B, Yu, J, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2010-01-05 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of holo and Cu-deficient Cu/Zn-SOD from the silkworm Bombyx mori and the implications in amyotrophic lateral sclerosis
Proteins, 78, 2010
|
|
3LYX
 
 | | Crystal structure of the PAS domain of the protein CPS_1291 from Colwellia psychrerythraea. Northeast Structural Genomics Consortium target id CsR222B | | Descriptor: | Sensory box/GGDEF domain protein | | Authors: | Seetharaman, J, Chen, Y, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-02-28 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the PAS domain of the protein CPS_1291 from Colwellia psychrerythraea. Northeast Structural Genomics Consortium target id CsR222B
To be Published
|
|
3MI6
 
 | | Crystal structure of the alpha-galactosidase from Lactobacillus brevis, Northeast Structural Genomics Consortium Target LbR11. | | Descriptor: | Alpha-galactosidase | | Authors: | Vorobiev, S, Chen, Y, Seetharaman, J, Belote, R, Sahdev, S, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-04-09 | | Release date: | 2010-04-28 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Crystal structure of the alpha-galactosidase from Lactobacillus brevis.
To be Published
|
|
3MER
 
 | | Crystal Structure of the methyltransferase Slr1183 from Synechocystis sp. PCC 6803, Northeast Structural Genomics Consortium Target SgR145 | | Descriptor: | Slr1183 protein | | Authors: | Seetharaman, J, Chen, Y, Forouhar, F, Rossi, P, Sahdev, S, Xiao, R, Ciccosanti, C, Foote, E.L, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Northeast Structural Genomics Consortium Target SgR145
To be Published
|
|
