9JAM
 
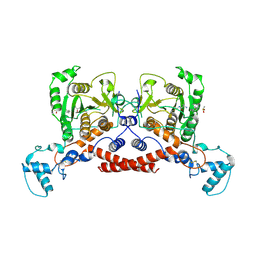 | | Cryo-EM structure of MPXV core protease in complex with compound A3 | | Descriptor: | A1ECK-DI8-ALA-AEM, Core protease I7 | | Authors: | Gao, Y, Xie, X, Lan, W, Wang, W, Yang, H. | | Deposit date: | 2024-08-25 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Substrate recognition and cleavage mechanism of the monkeypox virus core protease.
Nature, 2025
|
|
9JAQ
 
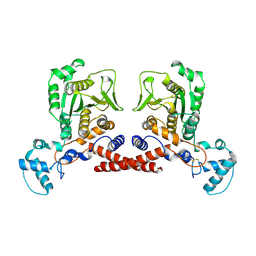 | | Cryo-EM structure of MPXV core protease in the apo-form | | Descriptor: | Core protease I7 | | Authors: | Lan, W, You, T, Li, D, Dong, X, Wang, H, Xu, J, Wang, W, Gao, Y, Yang, H. | | Deposit date: | 2024-08-25 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Substrate recognition and cleavage mechanism of the monkeypox virus core protease.
Nature, 2025
|
|
9KQV
 
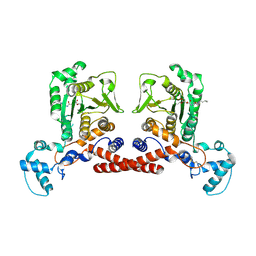 | | Cryo-EM structure of MPXV core protease in complex with aloxistatin(E64d) | | Descriptor: | Core protease I7, ethyl (3S)-3-hydroxy-4-({(2S)-4-methyl-1-[(3-methylbutyl)amino]-1-oxopentan-2-yl}amino)-4-oxobutanoate | | Authors: | Lan, W, You, T, Li, D, Dong, X, Wang, H, Xu, J, Wang, W, Gao, Y, Yang, H. | | Deposit date: | 2024-11-27 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Substrate recognition and cleavage mechanism of the monkeypox virus core protease.
Nature, 2025
|
|
9KR6
 
 | | Cryo-EM structure of MPXV core protease in complex with the substrate derivative I-G18 | | Descriptor: | Core protease I7, Core protein VP8 | | Authors: | Lan, W, You, T, Li, D, Dong, X, Wang, H, Xu, J, Wang, W, Gao, Y, Yang, H. | | Deposit date: | 2024-11-27 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Substrate recognition and cleavage mechanism of the monkeypox virus core protease.
Nature, 2025
|
|
4RC6
 
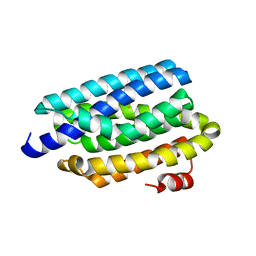 | | Crystal structure of cyanobacterial aldehyde-deformylating oxygenase 122F mutant | | Descriptor: | Aldehyde decarbonylase, FE (II) ION | | Authors: | Jia, C.J, Li, M, Li, J.J, Zhang, J.J, Zhang, H.M, Cao, P, Pan, X.W, Lu, X.F, Chang, W.R. | | Deposit date: | 2014-09-14 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the catalytic mechanism of aldehyde-deformylating oxygenases.
Protein Cell, 6, 2015
|
|
2CKX
 
 | |
6PWV
 
 | | Cryo-EM structure of MLL1 core complex bound to the nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
4RC8
 
 | |
4RC5
 
 | |
4QUW
 
 | |
5IIS
 
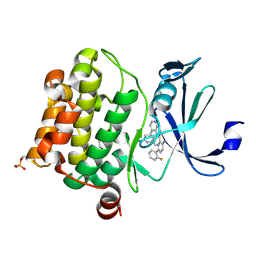 | | Design, synthesis and structure activity relationship of potent pan-PIM kinase inhibitors derived from the pyridyl-amide scaffold | | Descriptor: | 3-amino-N-(2'-amino-6'-methyl[4,4'-bipyridin]-3-yl)-6-(2-fluorophenyl)pyridine-2-carboxamide, DI(HYDROXYETHYL)ETHER, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C, Bussiere, D, Burger, M. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, synthesis and structure activity relationship of potent pan-PIM kinase inhibitors derived from the pyridyl carboxamide scaffold.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
4RC7
 
 | |
4RT4
 
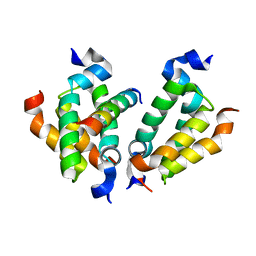 | | Crystal structure of Dpy30 complexed with Bre2 | | Descriptor: | Peptide from COMPASS component BRE2, Protein dpy-30 homolog | | Authors: | Zhang, H.M, Li, M, Chang, W.R. | | Deposit date: | 2014-11-12 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural implications of Dpy30 oligomerization for MLL/SET1 COMPASS H3K4 trimethylation
Protein Cell, 6, 2015
|
|
7WZ3
 
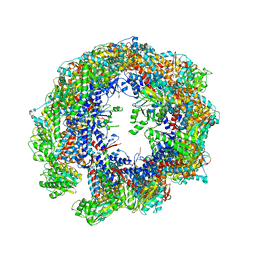 | | Cryo-EM structure of human TRiC-tubulin-S1 state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2022-02-16 | | Release date: | 2023-02-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Pathway and mechanism of tubulin folding mediated by TRiC/CCT along its ATPase cycle revealed using cryo-EM.
Commun Biol, 6, 2023
|
|
7X0V
 
 | | cryo-EM structure of human TRiC-ADP-AlFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2022-02-22 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Pathway and mechanism of tubulin folding mediated by TRiC/CCT along its ATPase cycle revealed using cryo-EM.
Commun Biol, 6, 2023
|
|
7X0A
 
 | | Cryo-EM structure of human TRiC-NPP state | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2022-02-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Pathway and mechanism of tubulin folding mediated by TRiC/CCT along its ATPase cycle revealed using cryo-EM.
Commun Biol, 6, 2023
|
|
7X3U
 
 | | cryo-EM structure of human TRiC-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2022-03-01 | | Release date: | 2023-06-07 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Pathway and mechanism of tubulin folding mediated by TRiC/CCT along its ATPase cycle revealed using cryo-EM.
Commun Biol, 6, 2023
|
|
7X3J
 
 | | Cryo-EM structure of human TRiC-tubulin-S2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2022-03-01 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Pathway and mechanism of tubulin folding mediated by TRiC/CCT along its ATPase cycle revealed using cryo-EM.
Commun Biol, 6, 2023
|
|
7X0S
 
 | | Human TRiC-tubulin-S3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2022-02-22 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Pathway and mechanism of tubulin folding mediated by TRiC/CCT along its ATPase cycle revealed using cryo-EM.
Commun Biol, 6, 2023
|
|
7X6Q
 
 | | cryo-EM structure of human TRiC-ATP-closed state | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2022-03-08 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Pathway and mechanism of tubulin folding mediated by TRiC/CCT along its ATPase cycle revealed using cryo-EM.
Commun Biol, 6, 2023
|
|
7X7Y
 
 | | Cryo-EM structure of Human TRiC-ATP-open state | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2022-03-10 | | Release date: | 2023-06-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Pathway and mechanism of tubulin folding mediated by TRiC/CCT along its ATPase cycle revealed using cryo-EM.
Commun Biol, 6, 2023
|
|
4RTA
 
 | |
7XHJ
 
 | | Crystal structure of RuvC from Deinococcus radiodurans | | Descriptor: | Crossover junction endodeoxyribonuclease RuvC | | Authors: | Qin, C, Zhao, Y. | | Deposit date: | 2022-04-08 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of the Holliday Junction Resolvase RuvC from Deinococcus radiodurans.
Microorganisms, 10, 2022
|
|
3BXV
 
 | |
7V26
 
 | | XG005-bound SARS-CoV-2 S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, XG005 Heavy chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2021-08-07 | | Release date: | 2021-10-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
