1CVR
 
 | |
6WEB
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
6WEC
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
2FTA
 
 | |
3E47
 
 | | Crystal Structure of the Yeast 20S Proteasome in Complex with Homobelactosin C | | Descriptor: | Proteasome component C1, Proteasome component C11, Proteasome component C5, ... | | Authors: | Groll, M, Larionov, O.V, de Meijere, A. | | Deposit date: | 2008-08-10 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibitor-binding mode of homobelactosin C to proteasomes: new insights into class I MHC ligand generation
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1ZIS
 
 | | Recombinant Lumazine synthase (hexagonal form) | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Lopez-Jaramillo, F.J. | | Deposit date: | 2005-04-27 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of recombinant Lumazine synthase (hexagonal form)
To be Published
|
|
2QXI
 
 | | High resolution structure of Human Kallikrein 7 in Complex with Suc-Ala-Ala-Pro-Phe-chloromethylketone | | Descriptor: | Kallikrein-7, N-(3-carboxypropanoyl)-L-alanyl-L-alanyl-N-[(2S,3S)-4-chloro-3-hydroxy-1-phenylbutan-2-yl]-L-prolinamide | | Authors: | Debela, M, Hess, P, Magdolen, V, Schechter, N.M, Bode, W, Goettig, P. | | Deposit date: | 2007-08-11 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Chymotryptic specificity determinants in the 1.0 A structure of the zinc-inhibited human tissue kallikrein 7.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4RCR
 
 | | STRUCTURE OF THE REACTION CENTER FROM RHODOBACTER SPHAEROIDES R-26 AND 2.4.1: PROTEIN-COFACTOR (BACTERIOCHLOROPHYLL, BACTERIOPHEOPHYTIN, AND CAROTENOID) INTERACTIONS | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Komiya, H, Yeates, T.O, Chirino, A.J, Rees, D.C, Allen, J.P, Feher, G. | | Deposit date: | 1991-09-09 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the reaction center from Rhodobacter sphaeroides R-26 and 2.4.1: protein-cofactor (bacteriochlorophyll, bacteriopheophytin, and carotenoid) interactions.
Proc.Natl.Acad.Sci.USA, 85, 1988
|
|
2CSC
 
 | |
1IAG
 
 | |
3ZI5
 
 | | Crystal STRUCTURE OF RESTRICTION ENDONUCLEASE BFII C-TERMINAL RECOGNITION DOMAIN IN COMPLEX WITH COGNATE DNA | | Descriptor: | 5'-D(*AP*GP*CP*AP*CP*TP*GP*GP*GP*TP*CP*GP)-3', 5'-D(*CP*GP*AP*CP*CP*CP*AP*GP*TP*GP*CP*TP)-3', RESTRICTION ENDONUCLEASE | | Authors: | Golovenko, D, Manakova, E, Zakrys, L, Zaremba, M, Sasnauskas, G, Grazulis, S, Siksnys, V. | | Deposit date: | 2013-01-03 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insight Into the Specificity of the B3 DNA-Binding Domains Provided by the Co-Crystal Structure of the C-Terminal Fragment of Bfii Restriction Enzyme
Nucleic Acids Res., 42, 2014
|
|
3OVO
 
 | |
4AIG
 
 | | ADAMALYSIN II WITH PHOSPHONATE INHIBITOR | | Descriptor: | ADAMALYSIN II, CALCIUM ION, N-[(FURAN-2-YL)CARBONYL]-(S)-LEUCYL-(R)-[1-AMINO-2(1H-INDOL-3-YL)ETHYL]-PHOSPHONIC ACID, ... | | Authors: | Pochetti, G, Mazza, F, Gavuzzo, E, Cirilli, M. | | Deposit date: | 1997-10-16 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2 angstrom X-ray structure of adamalysin II complexed with a peptide phosphonate inhibitor adopting a retro-binding mode.
FEBS Lett., 418, 1997
|
|
1GP1
 
 | |
1HAK
 
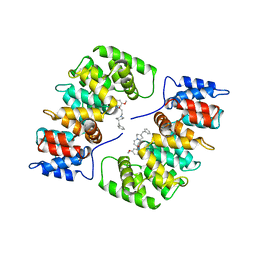 | | CRYSTAL STRUCTURE OF RECOMBINANT HUMAN PLACENTAL ANNEXIN V COMPLEXED WITH K-201 AS A CALCIUM CHANNEL ACTIVITY INHIBITOR | | Descriptor: | 4-[3-{1-(4-BENZYL)PIPERODINYL}PROPIONYL]-7-METHOXY-2,3,4,5-TERTRAHYDRO-1,4-BENZOTHIAZEPINE, ANNEXIN V | | Authors: | Ago, H, Inagaki, E, Miyano, M. | | Deposit date: | 1997-12-10 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of annexin V with its ligand K-201 as a calcium channel activity inhibitor.
J.Mol.Biol., 274, 1997
|
|
4BFH
 
 | |
3PRC
 
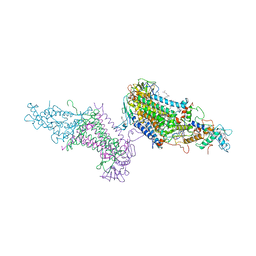 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (QB-DEPLETED) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Lancaster, C.R.D, Michel, H. | | Deposit date: | 1997-07-29 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The coupling of light-induced electron transfer and proton uptake as derived from crystal structures of reaction centres from Rhodopseudomonas viridis modified at the binding site of the secondary quinone, QB.
Structure, 5, 1997
|
|
1IAC
 
 | | REFINED 1.8 ANGSTROMS X-RAY CRYSTAL STRUCTURE OF ASTACIN, A ZINC-ENDOPEPTIDASE FROM THE CRAYFISH ASTACUS ASTACUS L. STRUCTURE DETERMINATION, REFINEMENT, MOLECULAR STRUCTURE AND COMPARISON WITH THERMOLYSIN | | Descriptor: | ASTACIN, MERCURY (II) ION | | Authors: | Gomis-Rueth, F.-X, Stoecker, W, Bode, W. | | Deposit date: | 1994-05-09 | | Release date: | 1994-08-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Refined 1.8 A X-ray crystal structure of astacin, a zinc-endopeptidase from the crayfish Astacus astacus L. Structure determination, refinement, molecular structure and comparison with thermolysin.
J.Mol.Biol., 229, 1993
|
|
2OW0
 
 | | MMP-9 active site mutant with iodine-labeled carboxylate inhibitor | | Descriptor: | CALCIUM ION, CHLORIDE ION, Matrix metalloproteinase-9 (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB), ... | | Authors: | Tochowicz, A, Bode, W, Maskos, K, Goettig, P. | | Deposit date: | 2007-02-15 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of MMP-9 Complexes with Five Inhibitors: Contribution of the Flexible Arg424 Side-chain to Selectivity.
J.Mol.Biol., 371, 2007
|
|
1NAG
 
 | | CREVICE-FORMING MUTANTS IN THE RIGID CORE OF BOVINE PANCREATIC TRYPSIN INHIBITOR: CRYSTAL STRUCTURES OF F22A, Y23A, N43G, AND F45A | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR, PHOSPHATE ION | | Authors: | Danishefsky, A.T, Wlodawer, A, Kim, K.-S, Tao, F, Woodward, C. | | Deposit date: | 1992-08-18 | | Release date: | 1993-10-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crevice-forming mutants in the rigid core of bovine pancreatic trypsin inhibitor: crystal structures of F22A, Y23A, N43G, and F45A.
Protein Sci., 2, 1993
|
|
1FC1
 
 | |
1GC2
 
 | | CRYSTAL STRUCTURE OF THE PYRIDOXAL-5'-PHOSPHATE DEPENDENT L-METHIONINE GAMMA-LYASE FROM PSEUDOMONAS PUTIDA | | Descriptor: | METHIONINE GAMMA-LYASE | | Authors: | Motoshima, H, Inagaki, K, Kumasaka, T, Furuichi, M, Inoue, H, Tamura, T, Esaki, N, Soda, K, Tanaka, N, Yamamoto, M, Tanaka, H. | | Deposit date: | 2000-07-06 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the pyridoxal 5'-phosphate dependent L-methionine gamma-lyase from Pseudomonas putida.
J.Biochem., 128, 2000
|
|
1GC0
 
 | | CRYSTAL STRUCTURE OF THE PYRIDOXAL-5'-PHOSPHATE DEPENDENT L-METHIONINE GAMMA-LYASE FROM PSEUDOMONAS PUTIDA | | Descriptor: | METHIONINE GAMMA-LYASE | | Authors: | Motoshima, H, Inagaki, K, Kumasaka, T, Furuichi, M, Inoue, H, Tamura, T, Esaki, N, Soda, K, Tanaka, N, Yamamoto, M, Tanaka, H. | | Deposit date: | 2000-07-06 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the pyridoxal 5'-phosphate dependent L-methionine gamma-lyase from Pseudomonas putida.
J.Biochem., 128, 2000
|
|
1GQF
 
 | |
2PSY
 
 | | Crystal Structure of Human Kallikrein 5 in complex with Leupeptin and Zinc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Kallikrein-5, LEUPEPTIN, ... | | Authors: | Debela, M, Bode, W, Goettig, P. | | Deposit date: | 2007-05-07 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of the zinc inhibition of human tissue kallikrein 5.
J.Mol.Biol., 373, 2007
|
|
