1L31
 
 | |
1L17
 
 | |
1L34
 
 | |
1L18
 
 | |
1L35
 
 | |
4WAK
 
 | |
4WAM
 
 | |
2FNO
 
 | |
6J6M
 
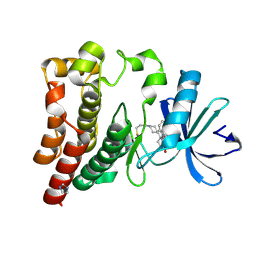 | | Co-crystal structure of BTK kinase domain with Zanubrutinib | | Descriptor: | (7S)-2-(4-phenoxyphenyl)-7-(1-propanoylpiperidin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, IMIDAZOLE, Tyrosine-protein kinase BTK | | Authors: | Zhou, X, Hong, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of Zanubrutinib (BGB-3111), a Novel, Potent, and Selective Covalent Inhibitor of Bruton's Tyrosine Kinase.
J.Med.Chem., 62, 2019
|
|
5NFT
 
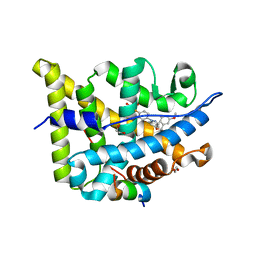 | | Glucocorticoid Receptor in complex with AZD5423 | | Descriptor: | 1,2-ETHANEDIOL, 2,2,2-tris(fluoranyl)-~{N}-[(1~{R},2~{S})-1-[1-(4-fluorophenyl)indazol-5-yl]oxy-1-(3-methoxyphenyl)propan-2-yl]ethanamide, Glucocorticoid receptor, ... | | Authors: | Edman, K, Wissler, L. | | Deposit date: | 2017-03-15 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Selective Nonsteroidal Glucocorticoid Receptor Modulators for the Inhaled Treatment of Pulmonary Diseases.
J. Med. Chem., 60, 2017
|
|
6Y14
 
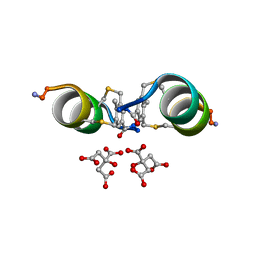 | |
6Y13
 
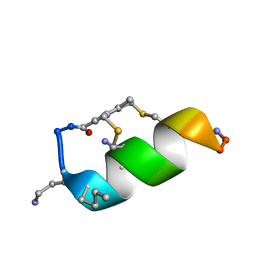 | |
6Y0U
 
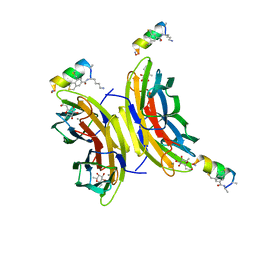 | | Fucosylated Bicyclic peptide bp71 bound to the fucose binding lectin LecB PA-IIL from Pseudomonas aeruginosa at 1.5 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, Fucose-binding lectin, ... | | Authors: | Baeriswyl, S, Stocker, A, Reymond, J.-L. | | Deposit date: | 2020-02-10 | | Release date: | 2021-02-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | A mixed chirality alpha-helix in a stapled bicyclic and a linear antimicrobial peptide revealed by X-ray crystallography.
Rsc Chem Biol, 2, 2021
|
|
6Y0V
 
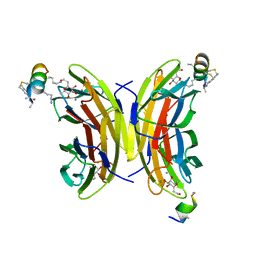 | | Fucosylated bicyclic peptide bp71 bound to the fucose binding lectin LecB PA-IIL from Pseudomonas aeruginosa at 1.7 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, Fucose-binding lectin, ... | | Authors: | Baeriswyl, S, Stocker, A, Reymond, J.-L. | | Deposit date: | 2020-02-10 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A mixed chirality alpha-helix in a stapled bicyclic and a linear antimicrobial peptide revealed by X-ray crystallography.
Rsc Chem Biol, 2, 2021
|
|
4V7O
 
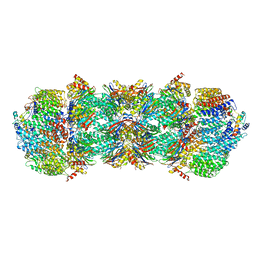 | | Proteasome Activator Complex | | Descriptor: | Proteasome activator BLM10, Proteasome component C1, Proteasome component C11, ... | | Authors: | Hill, C.P, Whitby, F.G. | | Deposit date: | 2009-12-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening.
Mol.Cell, 37, 2010
|
|
4WLR
 
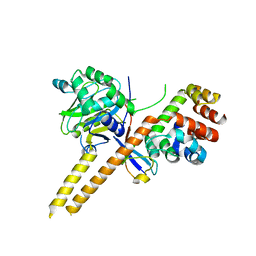 | | Crystal Structure of mUCH37-hRPN13 CTD-hUb complex | | Descriptor: | Polyubiquitin-B, Proteasomal ubiquitin receptor ADRM1, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Hemmis, C.W, Hill, C.P, VanderLinden, R, Whitby, F.G. | | Deposit date: | 2014-10-07 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural Basis for the Activation and Inhibition of the UCH37 Deubiquitylase.
Mol.Cell, 57, 2015
|
|
7RRB
 
 | | IDO1 IN COMPLEX WITH COMPOUND 9 | | Descriptor: | 3-[4-(6-cyclopropylpyridin-3-yl)phenyl]-N-(4-fluorophenyl)oxetane-3-carboxamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-08-09 | | Release date: | 2022-03-16 | | Last modified: | 2022-05-11 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Oxetane Promise Delivered: Discovery of Long-Acting IDO1 Inhibitors Suitable for Q3W Oral or Parenteral Dosing.
J.Med.Chem., 65, 2022
|
|
7RRC
 
 | | IDO1 IN COMPLEX WITH COMPOUND 14 | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(4-fluorophenyl)-3-{4-[4-(hydroxymethyl)-6-(trifluoromethyl)pyridin-3-yl]phenyl}oxetane-3-carboxamide | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-08-09 | | Release date: | 2022-03-16 | | Last modified: | 2022-05-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Oxetane Promise Delivered: Discovery of Long-Acting IDO1 Inhibitors Suitable for Q3W Oral or Parenteral Dosing.
J.Med.Chem., 65, 2022
|
|
7RRD
 
 | | IDO1 IN COMPLEX WITH COMPOUND S-1 | | Descriptor: | 3-[4-(1H-benzimidazol-2-yl)phenyl]-N-(4-fluorophenyl)oxetane-3-carboxamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Oxetane promise delivered: discovery of long acting IDO1 inhibitors suitable for Q3W oral or parenteral dosing
To Be Published
|
|
5NQR
 
 | | Potent inhibitors of NUDT5 silence hormone signaling in breast cancer | | Descriptor: | 8-(dimethylamino)-1,3-dimethyl-7-[[5-(3-methylphenyl)-1,3,4-oxadiazol-2-yl]methyl]purine-2,6-dione, ADP-sugar pyrophosphatase | | Authors: | Carter, M, Stenmark, P. | | Deposit date: | 2017-04-21 | | Release date: | 2018-01-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeted NUDT5 inhibitors block hormone signaling in breast cancer cells.
Nat Commun, 9, 2018
|
|
6Y1S
 
 | |
6WPE
 
 | | HUMAN IDO1 IN COMPLEX WITH COMPOUND 4 | | Descriptor: | 4-chloro-N-{[1-(3-chlorobenzene-1-carbonyl)-1,2,3,4-tetrahydroquinolin-6-yl]methyl}benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A, Lammens, A. | | Deposit date: | 2020-04-27 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Carbamate and N -Pyrimidine Mitigate Amide Hydrolysis: Structure-Based Drug Design of Tetrahydroquinoline IDO1 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
6IDZ
 
 | | Crystal structure of H7 hemagglutinin mutant H7-SVTQ ( A138S, P221T, L226Q) with 3'SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-12 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.707 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
6IDA
 
 | | Crystal structure of H7 hemagglutinin mutant H7-SVTQ ( A138S, P221T, L226Q) from the influenza virus A/Anhui/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-09 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
6ID8
 
 | | Crystal structure of H7 hemagglutinin mutant H7-SVTL ( A138S, P221T) from the influenza virus A/Anhui/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-09 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
