2IMJ
 
 | | X-ray Crystal Structure of Protein PFL_3262 from Pseudomonas fluorescens. Northeast Structural Genomics Consortium Target PlR14. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Hypothetical protein DUF1348 | | Authors: | Zhou, W, Forouhar, F, Seetharaman, J, Chen, C.X, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-10-04 | | Release date: | 2006-10-24 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the hypothetical protein (DUF1348) from Pseudomonas fluorescens, Northeast Structural Genomics target PlR14.
TO BE PUBLISHED
|
|
2IN5
 
 | | Crystal Structure of the hypothetical lipoprotein YmcC from Escherichia coli (K12), Northeast Structural Genomics target ER552. | | Descriptor: | Hypothetical lipoprotein ymcC | | Authors: | Zhou, W, Forouhar, F, Seetharaman, J, Chen, C.X, Cunningham, K, Ma, L.-C, Janjua, H, Xiao, R, Baran, M.C, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-10-05 | | Release date: | 2006-11-07 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the hypothetical lipoprotein YmcC from Escherichia coli (K12), Northeast Structural Genomics target ER552.
TO BE PUBLISHED
|
|
1T3G
 
 | |
2IQI
 
 | | Crystal structure of protein XCC0632 from Xanthomonas campestris, Pfam DUF330 | | Descriptor: | Hypothetical protein XCC0632 | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Mckenzie, C, Pelletier, L, Wasserman, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-13 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of hypothetical protein XCC0632 from Xanthomonas campestris pv. campestris
To be Published
|
|
1T7Z
 
 | | Zn-alpha-2-glycoprotein; baculo-ZAG no PEG, no glycerol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Zinc-alpha-2-glycoprotein | | Authors: | Delker, S.L, West Jr, A.P, McDermott, L, Kennedy, M.W, Bjorkman, P.J. | | Deposit date: | 2004-05-11 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic studies of ligand binding by Zn-alpha2-glycoprotein.
J.Struct.Biol., 148, 2004
|
|
1T9F
 
 | | Structural genomics of Caenorhabditis elegans: Structure of a protein with unknown function | | Descriptor: | MALONATE ION, protein 1d10 | | Authors: | Symersky, J, Li, S, Bunzel, R, Schormann, N, Luo, D, Huang, W.Y, Qiu, S, Gray, R, Zhang, Y, Arabashi, A, Lu, S, Luan, C.H, Tsao, J, DeLucas, L, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-16 | | Release date: | 2004-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: Structure of a protein with unknown function.
To be Published
|
|
1T9N
 
 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, SULFATE ION, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J, Tu, C.K, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-05-18 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Characterization of Active-Site Histidine as a Proton Shuttle in Catalysis
by Human Carbonic Anhydrase II
Biochemistry, 44, 2005
|
|
2J13
 
 | | Structure of a family 4 carbohydrate esterase from Bacillus anthracis | | Descriptor: | ACETATE ION, CACODYLATE ION, POLYSACCHARIDE DEACETYLASE, ... | | Authors: | Gloster, T.M, Oberbarnscheidt, L, Taylor, E.J, Davies, G.J. | | Deposit date: | 2006-08-08 | | Release date: | 2006-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a Carbohydrate Esterase from Bacillus Anthracis.
Proteins: Struct., Funct., Bioinf., 66, 2007
|
|
2IEI
 
 | | Crystal structure of rabbit muscle glycogen phosphorylase in complex with 3,4-dihydro-2-quinolone | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, (S)-2-CHLORO-N-(1-(2-(2-HYDROXYETHYLAMINO)-2-OXOETHYL)-2-OXO-1,2,3,4-TETRAHYDROQUINOLIN-3-YL)-6H-THIENO[2,3-B]PYRROLE-5-CARBOXAMIDE, Glycogen phosphorylase, ... | | Authors: | Birch, A.M, Kenny, P.W, Oikonomakos, N.G, Otterbein, L, Schofield, P, Whittamore, P.R.O, Whalley, D.P, Rowsell, S, Pauptit, R, Pannifer, A, Breed, J, Minshull, C. | | Deposit date: | 2006-09-19 | | Release date: | 2006-12-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Development of potent, orally active 1-substituted-3,4-dihydro-2-quinolone glycogen phosphorylase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
1TF0
 
 | | Crystal structure of the GA module complexed with human serum albumin | | Descriptor: | CITRIC ACID, DECANOIC ACID, Peptostreptococcal albumin-binding protein, ... | | Authors: | Lejon, S, Svensson, S, Bjorck, L, Frick, I.-M, Wikstrom, M. | | Deposit date: | 2004-05-26 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and biological implications of a bacterial albumin binding module in complex with human serum albumin
J.Biol.Chem., 279, 2004
|
|
2II8
 
 | | Anabaena sensory rhodopsin transducer | | Descriptor: | Anabaena sensory rhodopsin transducer protein | | Authors: | Vogeley, L, Sineshchekov, O.A, Trivedi, V.D, Spudich, E.N, Spudich, J.L, Luecke, H. | | Deposit date: | 2006-09-27 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the anabaena sensory rhodopsin transducer.
J.Mol.Biol., 367, 2007
|
|
1TJM
 
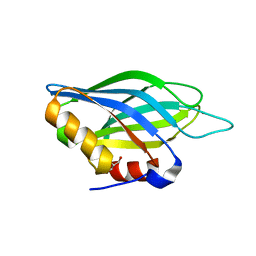 | | Crystallographic Identification of Sr2+ Coordination Site in Synaptotagmin I C2B Domain | | Descriptor: | GLYCEROL, STRONTIUM ION, Synaptotagmin I | | Authors: | Cheng, Y, Sequeira, S.M, Malinina, L, Tereshko, V, Sollner, T.H, Patel, D.J. | | Deposit date: | 2004-06-06 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystallographic identification of Ca2+ and Sr2+ coordination sites in synaptotagmin I C2B domain
Protein Sci., 13, 2004
|
|
2IPG
 
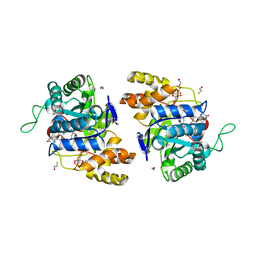 | | Crystal structure of 17alpha-hydroxysteroid dehydrogenase mutant K31A in complex with NADP+ and epi-testosterone | | Descriptor: | (10ALPHA,13ALPHA,14BETA,17ALPHA)-17-HYDROXYANDROST-4-EN-3-ONE, 1,2-ETHANEDIOL, 3(17)alpha-hydroxysteroid dehydrogenase, ... | | Authors: | Faucher, F, Cantin, L, Pereira de Jesus-Tran, K, Lemieux, M, Luu-the, V, Labrie, F, Breton, R. | | Deposit date: | 2006-10-12 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mouse 17alpha-Hydroxysteroid Dehydrogenase (AKR1C21) Binds Steroids Differently from other Aldo-keto Reductases: Identification and Characterization of Amino Acid Residues Critical for Substrate Binding.
J.Mol.Biol., 369, 2007
|
|
2IJR
 
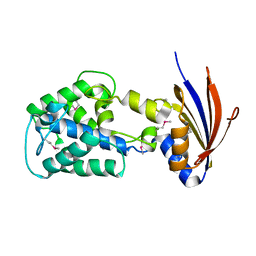 | | Crystal structure of a protein api92 from Yersinia pseudotuberculosis, Pfam DUF1281 | | Descriptor: | Hypothetical protein api92 | | Authors: | Jin, X, Min, T, Bonanno, J.B, Sauder, J.M, Wasserman, S, Smith, D, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-30 | | Release date: | 2006-10-31 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a hypothetical protein from Yersinia
pseudotuberculosis
To be Published
|
|
1TI1
 
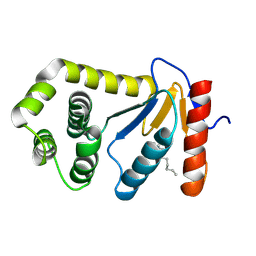 | | crystal structure of a mutant DsbA | | Descriptor: | DODECANE, Thiol:disulfide interchange protein dsbA | | Authors: | Kone, A, Serre, L. | | Deposit date: | 2004-06-02 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Intriguing conformation changes associated with the trans/cis isomerization of a prolyl residue in the active site of the DsbA C33A mutant.
J.Mol.Biol., 347, 2005
|
|
1TL4
 
 | | Solution structure of Cu(I) HAH1 | | Descriptor: | COPPER (I) ION, Copper transport protein ATOX1 | | Authors: | Anastassopoulou, I, Banci, L, Bertini, I, Cantini, F, Katsari, E, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-06-09 | | Release date: | 2004-10-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Apo and Copper(I)-Loaded Human Metallochaperone HAH1.
Biochemistry, 43, 2004
|
|
2IC1
 
 | | Crystal Structure of Human Cysteine Dioxygenase in Complex with Substrate Cysteine | | Descriptor: | CYSTEINE, Cysteine dioxygenase type 1, FE (II) ION | | Authors: | Ye, S, Wu, X, Wei, L, Tang, D, Sun, P, Rao, Z. | | Deposit date: | 2006-09-12 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An Insight into the Mechanism of Human Cysteine Dioxygenase: KEY ROLES OF THE THIOETHER-BONDED TYROSINE-CYSTEINE COFACTOR.
J.Biol.Chem., 282, 2007
|
|
2INQ
 
 | | Neutron Crystal Structure of Escherichia coli Dihydrofolate Reductase Bound to the Anti-cancer drug, Methotrexate | | Descriptor: | Dihydrofolate reductase, N-(4-{[(2,4-DIAMINOPTERIDIN-1-IUM-6-YL)METHYL](METHYL)AMINO}BENZOYL)-L-GLUTAMIC ACID | | Authors: | Bennett, B.C, Langan, P.A, Coates, L, Schoenborn, B, Dealwis, C.G. | | Deposit date: | 2006-10-08 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Neutron diffraction studies of Escherichia coli dihydrofolate reductase complexed with methotrexate.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1SWE
 
 | | APO-CORE-STREPTAVIDIN IN COMPLEX WITH BIOTIN AT PH 4.5 | | Descriptor: | BIOTIN, STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Klumb, L, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-03-04 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural studies of the streptavidin binding loop.
Protein Sci., 6, 1997
|
|
1SWD
 
 | | APO-CORE-STREPTAVIDIN IN COMPLEX WITH BIOTIN (TWO UNOCCUPIED BINDING SITES) AT PH 4.5 | | Descriptor: | BIOTIN, STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Klumb, L, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-03-04 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of the streptavidin binding loop.
Protein Sci., 6, 1997
|
|
1SWC
 
 | | APO-CORE-STREPTAVIDIN AT PH 4.5 | | Descriptor: | STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Klumb, L, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-03-04 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of the streptavidin binding loop.
Protein Sci., 6, 1997
|
|
1SWB
 
 | | APO-CORE-STREPTAVIDIN AT PH 7.5 | | Descriptor: | STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Klumb, L, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-03-04 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies of the streptavidin binding loop.
Protein Sci., 6, 1997
|
|
2J23
 
 | | Cross-reactivity and crystal structure of Malassezia sympodialis Thioredoxin (Mala s 13), a member of a new pan-allergen family | | Descriptor: | THIOREDOXIN | | Authors: | Limacher, A, Glaser, A.G, Meier, C, Scapozza, L, Crameri, R. | | Deposit date: | 2006-08-16 | | Release date: | 2006-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Cross-reactivity and 1.4-A crystal structure of Malassezia sympodialis thioredoxin (Mala s 13), a member of a new pan-allergen family.
J Immunol., 178, 2007
|
|
1SWA
 
 | | APO-CORE-STREPTAVIDIN AT PH 4.5 | | Descriptor: | STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Klumb, L, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-03-02 | | Release date: | 1998-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of the streptavidin binding loop.
Protein Sci., 6, 1997
|
|
4UZD
 
 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | AURORA KINASE A, ethyl (9S)-9-[3-(1H-benzimidazol-2-yloxy)phenyl]-8-oxo-4,5,6,7,8,9-hexahydro-2H-pyrrolo[3,4-b]quinoline-3-carboxylate | | Authors: | Pouzieux, S, Delarbre, L, Crenne, J.Y. | | Deposit date: | 2014-09-05 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Sar156497 an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
