1ZS7
 
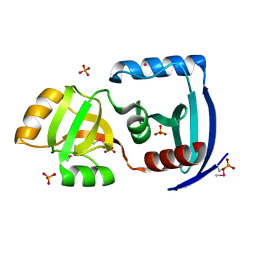 | | The structure of gene product APE0525 from Aeropyrum pernix | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, hypothetical protein APE0525 | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-23 | | Release date: | 2005-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of gene product APE0525 from Aeropyrum pernix
To be Published
|
|
2A9S
 
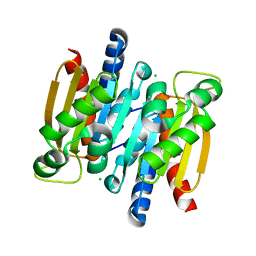 | | The crystal structure of competence/damage inducible protein CihA from Agrobacterium tumefaciens | | Descriptor: | CHLORIDE ION, competence/damage-inducible protein CinA | | Authors: | Lunin, V.V, Evdokimova, E, Kudritska, M, Kim, Y, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-07-12 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of competence/damage inducible protein CihA from Agrobacterium tumefaciens
To be Published
|
|
4XS5
 
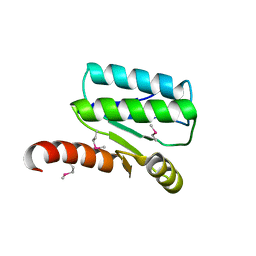 | | Crystal structure of Sulfate transporter/antisigma-factor antagonist STAS from Dyadobacter fermentans DSM 18053 | | Descriptor: | Sulfate transporter/antisigma-factor antagonist STAS | | Authors: | Chang, C, Cuff, M, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-21 | | Release date: | 2015-02-11 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of Sulfate transporter/antisigma-factor antagonist STAS from Dyadobacter fermentans DSM 18053
To Be Published
|
|
2R39
 
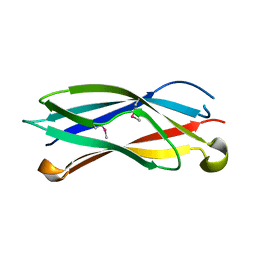 | |
2QSA
 
 | | Crystal structure of J-domain of DnaJ homolog dnj-2 precursor from C.elegans. | | Descriptor: | CHLORIDE ION, DnaJ homolog dnj-2 | | Authors: | Osipiuk, J, Mulligan, R, Gu, M, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray crystal structure of J-domain of DnaJ homolog dnj-2 precursor from C.elegans.
To be Published
|
|
4YCS
 
 | | Crystal structure of putative lipoprotein from Peptoclostridium difficile 630 (fragment) | | Descriptor: | ACETATE ION, GLYCEROL, SODIUM ION, ... | | Authors: | Michalska, K, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-20 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of putative lipoprotein from Peptoclostridium difficile 630 (fragment)
To Be Published
|
|
2QYB
 
 | |
2R2C
 
 | |
4YF1
 
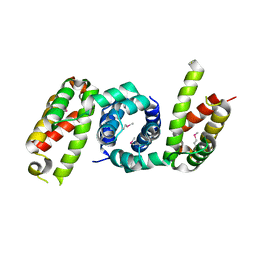 | | 1.85 angstrom crystal structure of lmo0812 from Listeria monocytogenes EGD-e | | Descriptor: | CITRATE ANION, Lmo0812 protein, SODIUM ION | | Authors: | Krishna, S.N, Light, S.H, Filippova, E.V, Minasov, G, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 angstrom crystal structure of lmo0812 from Listeria monocytogenes EGD-e
To Be Published
|
|
2QSW
 
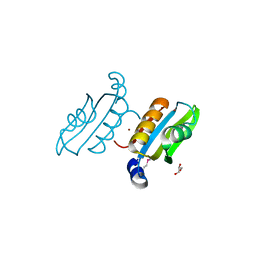 | | Crystal structure of C-terminal domain of ABC transporter / ATP-binding protein from Enterococcus faecalis | | Descriptor: | GLYCEROL, Methionine import ATP-binding protein metN 2, ZINC ION | | Authors: | Osipiuk, J, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-31 | | Release date: | 2007-08-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structure of C-terminal domain of ABC transporter / ATP-binding protein from Enterococcus faecalis.
To be Published
|
|
2QH9
 
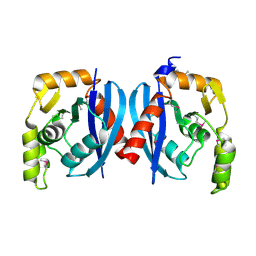 | |
2QHK
 
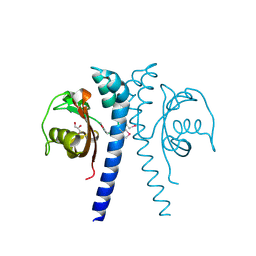 | |
2QZI
 
 | | The crystal structure of a conserved protein of unknown function from Streptococcus thermophilus LMG 18311. | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Uncharacterized protein | | Authors: | Tan, K, Zhou, M, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-16 | | Release date: | 2007-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of a conserved protein of unknown function from Streptococcus thermophilus LMG 18311.
To be Published
|
|
4Y7D
 
 | | Alpha/beta hydrolase fold protein from Nakamurella multipartita | | Descriptor: | Alpha/beta hydrolase fold protein, CHLORIDE ION, SODIUM ION | | Authors: | Cuff, M.E, OSIPIUK, J, Holowicki, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-14 | | Release date: | 2015-02-25 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Alpha/beta hydrolase fold protein from Nakamurella multipartita.
to be published
|
|
2QMX
 
 | | The crystal structure of L-Phe inhibited prephenate dehydratase from Chlorobium tepidum TLS | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PHENYLALANINE, ... | | Authors: | Tan, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-17 | | Release date: | 2007-08-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of open (R) and close (T) states of prephenate dehydratase (PDT) - implication of allosteric regulation by L-phenylalanine.
J.Struct.Biol., 162, 2008
|
|
2QIP
 
 | | Crystal structure of a protein of unknown function VPA0982 from Vibrio parahaemolyticus RIMD 2210633 | | Descriptor: | 1,2-ETHANEDIOL, Protein of unknown function VPA0982 | | Authors: | Tan, K, Duggan, E, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-05 | | Release date: | 2007-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The crystal structure of a protein of unknown function, VPA0982 from Vibrio parahaemolyticus RIMD 2210633.
To be Published
|
|
1T0B
 
 | | Structure of ThuA-like protein from Bacillus stearothermophilus | | Descriptor: | ThuA-like protein, ZINC ION | | Authors: | Borek, D, Chen, Y, Collart, F, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-04-08 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analysis of chemical interactions in ThuA-like protein from Bacillus Stearothermophilus
To be Published
|
|
1MKZ
 
 | | Crystal structure of MoaB protein at 1.6 A resolution. | | Descriptor: | ACETIC ACID, Molybdenum cofactor biosynthesis protein B, SULFATE ION | | Authors: | Sanishvili, R, Skarina, T, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-29 | | Release date: | 2003-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Escherichia coli MoaB suggests a probable role in molybdenum cofactor synthesis.
J.Biol.Chem., 279, 2004
|
|
2R2A
 
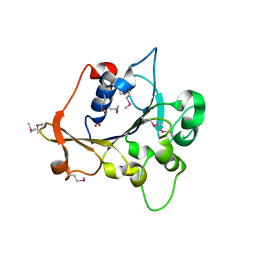 | | Crystal structure of N-terminal domain of zonular occludens toxin from Neisseria meningitidis | | Descriptor: | SULFATE ION, Uncharacterized protein | | Authors: | Osipiuk, J, Patterson, S, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-24 | | Release date: | 2007-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of N-terminal domain of zonular occludens toxin from Neisseria meningitidis.
To be Published
|
|
1T33
 
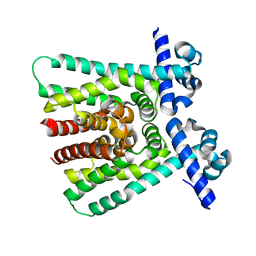 | |
1SS4
 
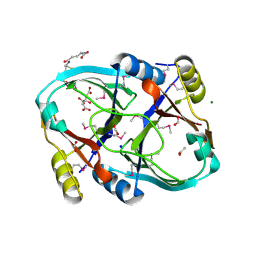 | |
2QSX
 
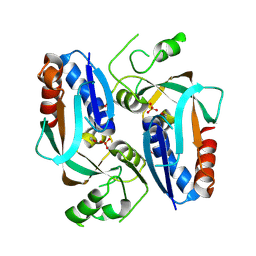 | | Crystal structure of putative transcriptional regulator LysR From Vibrio parahaemolyticus | | Descriptor: | Putative transcriptional regulator, LysR family, SULFATE ION | | Authors: | Wu, R, Abdullah, J, Binkowski, T.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-31 | | Release date: | 2007-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The Crystal Structure of Putative Transcriptional Regulator LysR From Vibrio parahaemolyticus.
To be Published
|
|
2QLC
 
 | |
2QHQ
 
 | | Crystal structure of unknown function protein VPA0580 | | Descriptor: | ACETATE ION, Unknown function protein VPA0580 | | Authors: | Chang, C, Kim, Y, Volkart, L, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-02 | | Release date: | 2007-07-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of unknown function protein VPA0580.
To be Published
|
|
2QSB
 
 | |
