6G80
 
 | |
3ER3
 
 | | The active site of aspartic proteinases | | Descriptor: | 6-ammonio-N-[(2R,4R,5R)-5-{[N-(tert-butoxycarbonyl)-L-phenylalanyl-3-(1H-imidazol-3-ium-4-yl)-L-alanyl]amino}-6-cyclohexyl-4-hydroxy-2-(2-methylpropyl)hexanoyl]-L-norleucylphenylalanine, ENDOTHIAPEPSIN | | Authors: | Al-Karadaghi, S, Cooper, J.B, Veerapandian, B, Hoover, D, Blundell, T.L. | | Deposit date: | 1991-01-02 | | Release date: | 1991-04-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Active Site of Aspartic Proteinases
FEBS Lett., 174, 1984
|
|
6GS8
 
 | | Crystal structure of SmbA in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, Uncharacterized protein | | Authors: | Dubey, B.N, Schirmer, T. | | Deposit date: | 2018-06-13 | | Release date: | 2019-07-31 | | Last modified: | 2022-07-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Reciprocal growth control by competitive binding of nucleotide second messengers to a metabolic switch in Caulobacter crescentus
Nat Microbiol, 2021
|
|
6GTH
 
 | | Serial Femtosecond Crystallography at Megahertz pulse rates | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Wiedorn, M, Oberthuer, D, Werner, N, Schubert, R, White, T.A, Mancuso, A, Perbandt, M, Betzel, C, Barty, A, Chapman, H. | | Deposit date: | 2018-06-18 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Megahertz serial crystallography.
Nat Commun, 9, 2018
|
|
6GTM
 
 | | Crystal structure of SmbA in complex with ppGpp. | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, SmbA | | Authors: | Dubey, B.N, Schirmer, T. | | Deposit date: | 2018-06-18 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Reciprocal growth control by competitive binding of nucleotide second messengers to a metabolic switch in Caulobacter crescentus
Nat Microbiol, 2021
|
|
6GL6
 
 | | apo [FeFe]-hydrogenase HydA1 from Chlamydomonas reinhardtii, variant C377H | | Descriptor: | CHLORIDE ION, Fe-hydrogenase, IRON/SULFUR CLUSTER, ... | | Authors: | Kertess, L, Happe, T, Hofmann, E. | | Deposit date: | 2018-05-23 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | His-Ligation to the [4Fe-4S] Subcluster Tunes the Catalytic Bias of [FeFe] Hydrogenase.
J.Am.Chem.Soc., 141, 2019
|
|
6GZ7
 
 | | Polyamide - DNA complex NMR structure | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*GP*TP*AP*CP*AP*TP*CP*G)-3'), dimethyl-[3-[3-[[1-methyl-4-[[1-methyl-4-[[1-methyl-4-[[1-methyl-4-[4-[[1-methyl-4-[[1-methyl-4-[[1-methyl-4-[(1-propan-2-ylimidazol-2-yl)carbonylamino]pyrrol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]butanoylamino]imidazol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]propanoylamino]propyl]azanium | | Authors: | Aman, K. | | Deposit date: | 2018-07-03 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Kinetic Profiling of Allosteric Modulation of Duplex DNA Induced by DNA-Binding Polyamide Analogues.
Chemistry, 25, 2019
|
|
5MG0
 
 | | Structure of PAS-GAF fragment of Deinococcus phytochrome by serial femtosecond crystallography | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, ... | | Authors: | Burgie, E.S, Fuller, F.D, Gul, S, Miller, M.D, Young, I.D, Brewster, A.S, Clinger, J, Aller, P, Braeuer, P, Hutchison, C, Alonso-Mori, R, Kern, J, Yachandra, V.K, Yano, J, Sauter, N.K, Phillips Jr, G.N, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers.
Nat. Methods, 14, 2017
|
|
5MU7
 
 | | Crystal Structure of the beta/delta-COPI Core Complex | | Descriptor: | Coatomer subunit beta, Coatomer subunit delta-like protein | | Authors: | Kopp, J, Aderhold, P, Wieland, F, Sinning, I. | | Deposit date: | 2017-01-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
6AGT
 
 | | Crystal structure of PfKRS complexed with chromone inhibitor | | Descriptor: | COBALT (II) ION, FORMIC ACID, LYSINE, ... | | Authors: | Yogavel, M, Sharma, A, Sharma, A, Baragana, B, Walpole, C. | | Deposit date: | 2018-08-14 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Lysyl-tRNA synthetase as a drug target in malaria and cryptosporidiosis.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6ALN
 
 | | VioC L-arginine hydroxylase bound to Fe(II), 3S-hydroxy-L-arginine, and 2OG | | Descriptor: | (2S,3S)-3-HYDROXYARGININE, 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent L-arginine hydroxylase, ... | | Authors: | Mitchell, A.J, Dunham, N.P, Boal, A.K. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Visualizing the Reaction Cycle in an Iron(II)- and 2-(Oxo)-glutarate-Dependent Hydroxylase.
J. Am. Chem. Soc., 139, 2017
|
|
6ALO
 
 | | VioC L-arginine hydroxylase bound to Fe(II), L-arginine, and a peroxysuccinate intermediate | | Descriptor: | 4-peroxy-4-oxobutanoic acid, ARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, ... | | Authors: | Dunham, N.P, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Visualizing the Reaction Cycle in an Iron(II)- and 2-(Oxo)-glutarate-Dependent Hydroxylase.
J. Am. Chem. Soc., 139, 2017
|
|
6GZP
 
 | | Llama nanobody PorM_02 structure determined at room temperature by in-situ diffraction in ChipX microfluidic device | | Descriptor: | Nanobody | | Authors: | Roche, J, Gaubert, A, Desmyter, A, De Wijn, R, Sauter, C, Roussel, A. | | Deposit date: | 2018-07-04 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
5MG1
 
 | | Structure of the photosensory module of Deinococcus phytochrome by serial femtosecond X-ray crystallography | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Burgie, E.S, Fuller, F.D, Gul, S, Young, I.D, Brewster, A.S, Clinger, J, Andi, B, Stan, C, Allaire, M, Nelsen, S, Alonso-Mori, R, Phillips Jr, G.N, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers.
Nat. Methods, 14, 2017
|
|
6A7K
 
 | | X-ray structure of NdhS from T. elongatus | | Descriptor: | ACETIC ACID, Tlr0636 protein | | Authors: | Umeno, K, Misumi, Y, Tanaka, H, Kurisu, G. | | Deposit date: | 2018-07-03 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural adaptations of photosynthetic complex I enable ferredoxin-dependent electron transfer.
Science, 363, 2019
|
|
6H40
 
 | | High resolution structure of MeT1 from Mycobacterium hassiacum in complex with 3-methoxy-1,2-propanediol. | | Descriptor: | (2~{S})-3-methoxypropane-1,2-diol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pereira, P.J.B, Ripoll-Rozada, J. | | Deposit date: | 2018-07-20 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.053 Å) | | Cite: | Biosynthesis of mycobacterial methylmannose polysaccharides requires a unique 1-O-methyltransferase specific for 3-O-methylated mannosides.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5M8E
 
 | | Crystal structure of a GH43 arabonofuranosidase from Weissella sp. strain 142 | | Descriptor: | Alpha-N-arabinofuranosidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Linares-Pasten, J, Logan, D.T, Nordberg Karlsson, E. | | Deposit date: | 2016-10-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures and functional studies of two GH43 arabinofuranosidases from Weissella sp. strain 142 and Lactobacillus brevis.
FEBS J., 284, 2017
|
|
6HCN
 
 | | Adenovirus Type 5 Fiber Knob protein at 1.49A resolution | | Descriptor: | 1,2-ETHANEDIOL, Fiber protein, MAGNESIUM ION | | Authors: | Rizkallah, P.J, Parker, A.L, Baker, A.T. | | Deposit date: | 2018-08-15 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Diversity within the adenovirus fiber knob hypervariable loops influences primary receptor interactions.
Nat Commun, 10, 2019
|
|
6HHO
 
 | | Crystal structure of RIP1 kinase in complex with GSK547 | | Descriptor: | 6-[4-[(5~{S})-5-[3,5-bis(fluoranyl)phenyl]pyrazolidin-1-yl]carbonylpiperidin-1-yl]pyrimidine-4-carbonitrile, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Thorpe, J.H, Harris, P.A. | | Deposit date: | 2018-08-28 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | RIP1 Kinase Drives Macrophage-Mediated Adaptive Immune Tolerance in Pancreatic Cancer.
Cancer Cell, 34, 2018
|
|
6ALM
 
 | | VioC L-arginine hydroxylase bound to Fe(II), L-arginine, and 2-OXO-GLUTARIC ACID | | Descriptor: | 2-OXOGLUTARIC ACID, ARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, ... | | Authors: | Dunham, N.P, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Visualizing the Reaction Cycle in an Iron(II)- and 2-(Oxo)-glutarate-Dependent Hydroxylase.
J. Am. Chem. Soc., 139, 2017
|
|
6ALQ
 
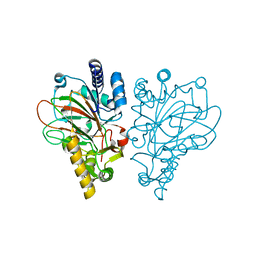 | | VioC L-arginine hydroxylase bound to Fe(II), L-arginine, and succinate | | Descriptor: | ARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, FE (II) ION, ... | | Authors: | Dunham, N.P, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Visualizing the Reaction Cycle in an Iron(II)- and 2-(Oxo)-glutarate-Dependent Hydroxylase.
J. Am. Chem. Soc., 139, 2017
|
|
6HFX
 
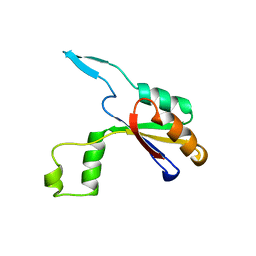 | |
6ALP
 
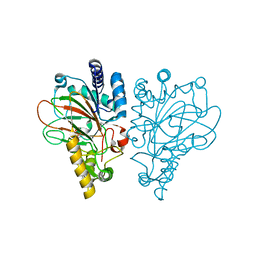 | | VioC L-arginine hydroxylase bound to Fe(II), 3S-hydroxy-L-arginine, and succinate | | Descriptor: | (2S,3S)-3-HYDROXYARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, FE (II) ION, ... | | Authors: | Mitchell, A.J, Dunham, N.P, Boal, A.K. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Visualizing the Reaction Cycle in an Iron(II)- and 2-(Oxo)-glutarate-Dependent Hydroxylase.
J. Am. Chem. Soc., 139, 2017
|
|
5MXS
 
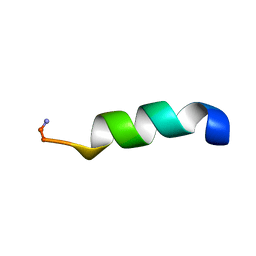 | |
6HCU
 
 | | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum bound to a difluoro cyclohexyl chromone ligand | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, HISTIDINE, ... | | Authors: | Tamjar, J, Robinson, D.A, Baragana, B, Norcross, N, Forte, B, Walpole, C, Gilbert, I.H. | | Deposit date: | 2018-08-16 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Lysyl-tRNA synthetase as a drug target in malaria and cryptosporidiosis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
