4FON
 
 | |
3D7E
 
 | |
5U8E
 
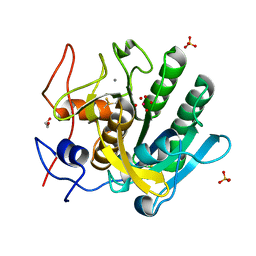
 | | Crystal Structure of substrate-free arginine kinase from spider Polybetes pythagoricus | | Descriptor: | SODIUM ION, arginine kinase | | Authors: | Lopez-Zavala, A.A, Garcia, C.F, Hernadez-Paredes, J, Sotelo-Mundo, R.R. | | Deposit date: | 2016-12-14 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Biochemical and structural characterization of a novel arginine kinase from the spider Polybetes pythagoricus.
PeerJ, 5, 2017
|
|
6OH6
 
 | |
4CS5
 
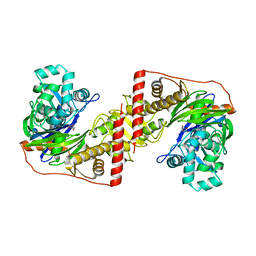
 | | Crystal Structure of PCNA from Litopenaeus vannamei | | Descriptor: | PROLIFERATING CELL NUCLEAR ANTIGEN | | Authors: | Carrasco-Miranda, J.S, Lopez-Zavala, A.A, De-La-Mora, E, Rudino-Pinera, E, Brieba, L.G, Sotelo-Mundo, R.R. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the Shrimp Proliferating Cell Nuclear Antigen: Structural Complementarity with Wssv DNA Polymerase Pip-Box.
Plos One, 9, 2014
|
|
4X3T
 
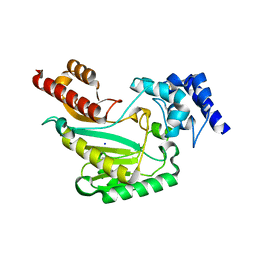
 | | Crystal structure of chromobox homolog 7 (CBX7) chromodomain with MS37452 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(2,3-dimethoxybenzoyl)piperazin-1-yl]-2-(3-methylphenoxy)ethanone, Chromobox protein homolog 7, ... | | Authors: | Ren, C, Jakoncic, J, Zhou, M.M. | | Deposit date: | 2014-12-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Small-Molecule Modulators of Methyl-Lysine Binding for the CBX7 Chromodomain.
Chem.Biol., 22, 2015
|
|
4B5L
 
 | |
4X3K
 
 | |
4X3U
 
 | | Crystal structure of chromobox homolog 7 (CBX7) chromodomain with Suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, Chromobox protein homolog 7 | | Authors: | Ren, C, Zhou, M.M. | | Deposit date: | 2014-12-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Small-Molecule Modulators of Methyl-Lysine Binding for the CBX7 Chromodomain.
Chem.Biol., 22, 2015
|
|
4X3S
 
 | | Crystal structure of chromobox homology 7 (CBX7) with SETDB1-1170me3 Peptide | | Descriptor: | CITRIC ACID, Chromobox protein homolog 7, FE (III) ION, ... | | Authors: | Ren, C, Plotnikov, A.N, Zhou, M.M. | | Deposit date: | 2014-12-01 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small-Molecule Modulators of Methyl-Lysine Binding for the CBX7 Chromodomain.
Chem.Biol., 22, 2015
|
|
3VFK
 
 | | The structure of monodechloro-teicoplanin in complex with its ligand, using ubiquitin as a ligand carrier | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose, 8-METHYLNONANOIC ACID, ... | | Authors: | Economou, N.J, Weeks, S.D, Grasty, K.C, Loll, P.J. | | Deposit date: | 2012-01-09 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the complex between teicoplanin and a bacterial cell-wall peptide: use of a carrier-protein approach.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6OH7
 
 | |
6OH8
 
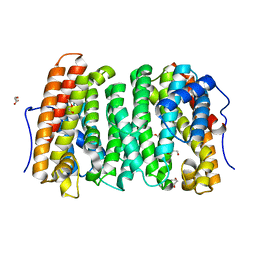 | |
2FBO
 
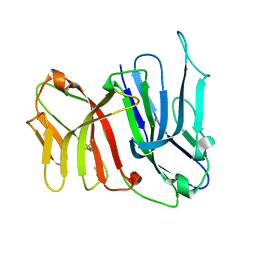 | | Crystal Structure of the Two Tandem V-type Regions of VCBP3 (v-region-containing chitin binding protein) to 1.85 A | | Descriptor: | variable region-containing chitin-binding protein 3 | | Authors: | Hernandez Prada, J.A, Haire, R.N, Jakoncic, J, Cannon, J.P, Litman, G.W, Ostrov, D.A. | | Deposit date: | 2005-12-09 | | Release date: | 2006-10-17 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ancient evolutionary origin of diversified variable regions demonstrated by crystal structures of an immune-type receptor in amphioxus
Nat.Immunol., 7, 2006
|
|
2YAQ
 
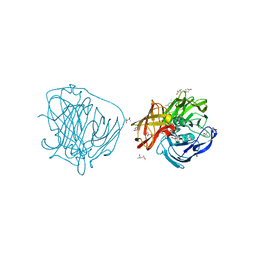 | | X-ray induced reduction of laccase from Thermus thermophilus HB27 (75. 0-87.5 percent dose) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Serrano-Posada, H, Rudino-Pinera, E. | | Deposit date: | 2011-02-24 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2YAM
 
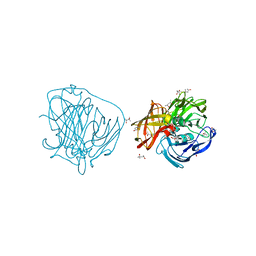 | | X-ray induced reduction of laccase from Thermus thermophilus HB27 (37. 5-50.0 percent dose) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Serrano-Posada, H, Rudino-Pinera, E. | | Deposit date: | 2011-02-23 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2YAH
 
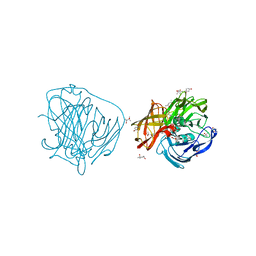 | | X-ray induced reduction of laccase from Thermus thermophilus HB27 (25. 0-37.5 percent dose) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Serrano-Posada, H, Rudino-Pinera, E. | | Deposit date: | 2011-02-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2YAE
 
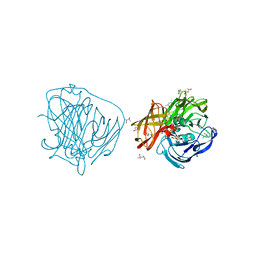 | | X-ray induced reduction of laccase from Thermus thermophilus HB27(0.0- 12.5 percent dose) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Serrano-Posada, H, Rudino-Pinera, E. | | Deposit date: | 2011-02-21 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2YAO
 
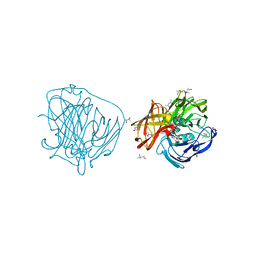 | | X-ray induced reduction of laccase from Thermus thermophilus HB27 (50. 0-62.5 percent dose) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Serrano-Posada, H, Rudino-Pinera, E. | | Deposit date: | 2011-02-23 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2YAR
 
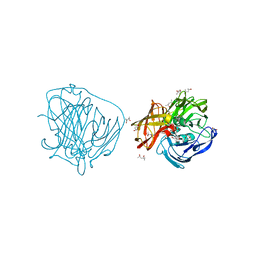 | | X-ray induced reduction of laccase from Thermus thermophilus HB27 (87. 5-100.0 percent dose) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Serrano-Posada, H, Rudino-Pinera, E. | | Deposit date: | 2011-02-24 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2YAF
 
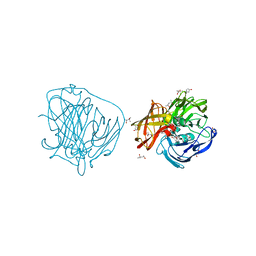 | | X-ray induced reduction of laccase from Thermus thermophilus HB27 (12. 5-25.0 percent dose) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Serrano-Posada, H, Rudino-Pinera, E. | | Deposit date: | 2011-02-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2YAP
 
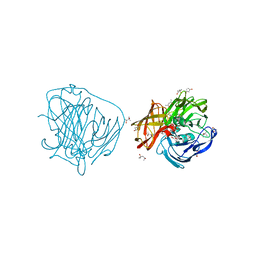 | | X-ray induced reduction of laccase from Thermus thermophilus HB27 (62. 5-75.0 percent dose) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Serrano-Posada, H, Rudino-Pinera, E. | | Deposit date: | 2011-02-23 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3VFJ
 
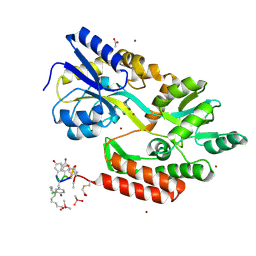 | | The structure of monodechloro-teicoplanin in complex with its ligand, using MBP as a ligand carrier | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose, 8-METHYLNONANOIC ACID, ... | | Authors: | Economou, N.J, Weeks, S.D, Grasty, K.C, Loll, P.J. | | Deposit date: | 2012-01-09 | | Release date: | 2013-01-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the complex between teicoplanin and a bacterial cell-wall peptide: use of a carrier-protein approach.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5KEJ
 
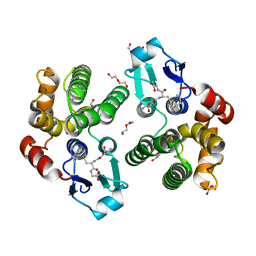 | | Crystallographic structure of the Tau class glutathione S-transferase MiGSTU in complex with S-hexyl-glutathione | | Descriptor: | DI(HYDROXYETHYL)ETHER, S-HEXYLGLUTATHIONE, Tau class glutathione S-transferase | | Authors: | Valenzuela-Chavira, I, Serrano-Posada, H, Lopez-Zavala, A, Hernandez-Paredes, J, Sotelo-Mundo, R. | | Deposit date: | 2016-06-09 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Insights into ligand binding to a glutathione S-transferase from mango: Structure, thermodynamics and kinetics.
Biochimie, 135, 2017
|
|
5K5K
 
 | | Crystal structure of laccasse from Thermus thermophilus HB27 (ascorbic acid 10 min) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Laccase | | Authors: | Diaz-Vilchis, A, Ruiz-Arellano, R.R, Rosas-Benitez, E, Rudino-Pinera, E. | | Deposit date: | 2016-05-23 | | Release date: | 2017-06-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Preserving metallic sites affected by radiation damage: the CuT2 case in Thermus thermophilus multicopper oxidase
To be Published
|
|
