7SGX
 
 | | Crystal Structure of Turtle Cadherin-23 EC1-2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cadherin 23, ... | | Authors: | Nisler, C.R, Sotomayor, M. | | Deposit date: | 2021-10-07 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Interpreting the Evolutionary Echoes of a Protein Complex Essential for Inner-Ear Mechanosensation.
Mol.Biol.Evol., 40, 2023
|
|
7SKH
 
 | |
6MFO
 
 | |
6MGA
 
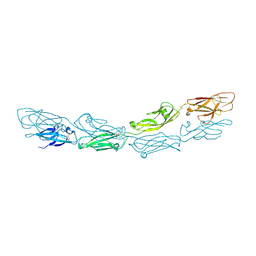 | |
2YMB
 
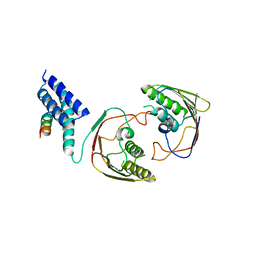 | | Structures of MITD1 | | Descriptor: | CHARGED MULTIVESICULAR BODY PROTEIN 1A, MIT DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Hadders, M.A, Agromayor, M, Obita, T, Perisic, O, Caballe, A, Kloc, M, Lamers, M.H, Williams, R.L, Martin-Serrano, J. | | Deposit date: | 2012-10-08 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | Escrt-III Binding Protein Mitd1 is Involved in Cytokinesis and Has an Unanticipated Pld Fold that Binds Membranes.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6N22
 
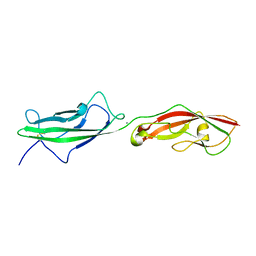 | |
6N2E
 
 | |
7SUU
 
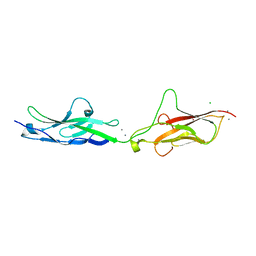 | |
7T80
 
 | |
7SZ8
 
 | |
7T3S
 
 | |
6ULM
 
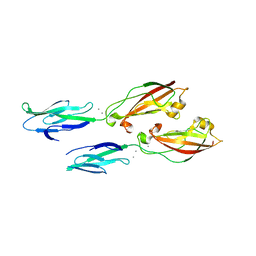 | | Crystal structure of human cadherin 17 EC1-2 | | Descriptor: | CALCIUM ION, Cadherin-17 | | Authors: | Gray, M.E, Sotomayor, M. | | Deposit date: | 2019-10-08 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the nonclassical cadherin-17 N-terminus and implications for its adhesive binding mechanism.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
6NW0
 
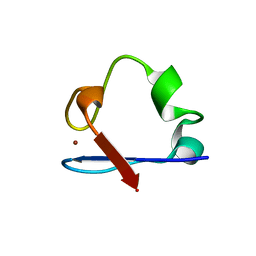 | | Crystal Structure Desulfovibrio desulfuricans Nickel-Substituted Rubredoxin | | Descriptor: | NICKEL (II) ION, Rubredoxin | | Authors: | Slater, J.W, Marguet, S.C, Gray, M.E, Sotomayor, M, Shafaat, H.S. | | Deposit date: | 2019-02-05 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Power of the Secondary Sphere: Modulating Hydrogenase Activity in Nickel-Substituted Rubredoxin
Acs Catalysis, 2019
|
|
6TPB
 
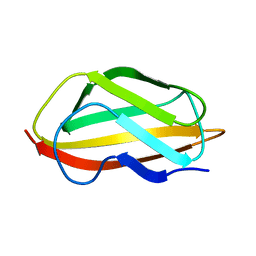 | | NMR structure of the apo-form of Pseudomonas fluorescens CopC | | Descriptor: | Putative copper resistance protein | | Authors: | Persson, K.C, Mayzel, M, Karlsson, B.G, Peciulyte, A, Olsson, L, Wittung Stafshede, P, Salomon Johansen, K, Horvath, I. | | Deposit date: | 2019-12-13 | | Release date: | 2021-01-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Pseudomonas fluorescens CopC
To Be Published
|
|
6NW1
 
 | | Crystal Structure Desulfovibrio desulfuricans Nickel-Substituted Rubredoxin V37N | | Descriptor: | NICKEL (II) ION, Rubredoxin | | Authors: | Slater, J.W, Marguet, S.C, Gray, M.E, Sotomayor, M, Shafaat, H.S. | | Deposit date: | 2019-02-05 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Power of the Secondary Sphere: Modulating Hydrogenase Activity in Nickel-Substituted Rubredoxin
Acs Catalysis, 2019
|
|
5I8D
 
 | |
5M3A
 
 | | Crystal structure of BRD4 BROMODOMAIN 1 IN COMPLEX WITH LIGAND 2 | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-6-(1-methyl-5-phenoxy-pyrazol-4-yl)-[1,2,4]triazolo[4,3-b]pyridazine, Bromodomain-containing protein 4 | | Authors: | Kessler, D, Mayer, M, Engelhardt, H, Wolkerstorfer, B, Geist, L. | | Deposit date: | 2016-10-14 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Direct NMR Probing of Hydration Shells of Protein Ligand Interfaces and Its Application to Drug Design.
J. Med. Chem., 60, 2017
|
|
5IU9
 
 | |
4A5Z
 
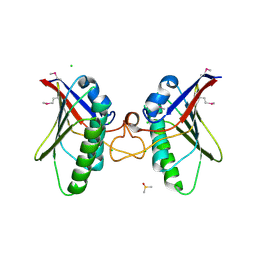 | | Structures of MITD1 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, MIT DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Hadders, M.A, Agromayor, M, Caballe, A, Obita, T, Perisic, O, Williams, R.L, Martin-Serrano, J. | | Deposit date: | 2011-10-30 | | Release date: | 2012-11-14 | | Last modified: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Escrt-III Binding Protein Mitd1 is Involved in Cytokinesis and Has an Unanticipated Pld Fold that Binds Membranes.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1A6A
 
 | | THE STRUCTURE OF AN INTERMEDIATE IN CLASS II MHC MATURATION: CLIP BOUND TO HLA-DR3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Ghosh, P, Amaya, M, Mellins, E, Wiley, D.C. | | Deposit date: | 1998-02-22 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structure of an intermediate in class II MHC maturation: CLIP bound to HLA-DR3.
Nature, 378, 1995
|
|
5M39
 
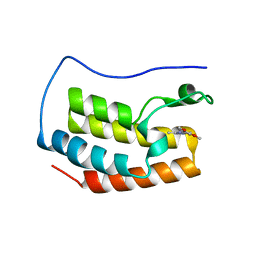 | | Crystal structure of BRD4 BROMODOMAIN 1 IN COMPLEX WITH LIGAND 1 | | Descriptor: | 6-(3,4-dimethoxyphenyl)-3-methyl-[1,2,4]triazolo[4,3-b]pyridazine, Bromodomain-containing protein 4 | | Authors: | Kessler, D, Mayer, M, Engelhardt, H, Wolkerstorfer, B, Geist, L. | | Deposit date: | 2016-10-14 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Direct NMR Probing of Hydration Shells of Protein Ligand Interfaces and Its Application to Drug Design.
J. Med. Chem., 60, 2017
|
|
4A5X
 
 | | Structures of MITD1 | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, CHARGED MULTIVESICULAR BODY PROTEIN 1A, GLYCEROL, ... | | Authors: | Hadders, M.A, Agromayor, M, Caballe, A, Obita, T, Perisic, O, Williams, R.L, Martin-Serrano, J. | | Deposit date: | 2011-10-28 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Escrt-III Binding Protein Mitd1 is Involved in Cytokinesis and Has an Unanticipated Pld Fold that Binds Membranes.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1FW9
 
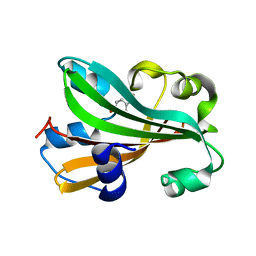 | | CHORISMATE LYASE WITH BOUND PRODUCT | | Descriptor: | CHORISMATE LYASE, P-HYDROXYBENZOIC ACID | | Authors: | Gallagher, D.T, Mayhew, M, Holden, M, Vilker, V, Howard, A. | | Deposit date: | 2000-09-22 | | Release date: | 2001-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structure of chorismate lyase shows a new fold and a tightly retained product.
Proteins, 44, 2001
|
|
1G1B
 
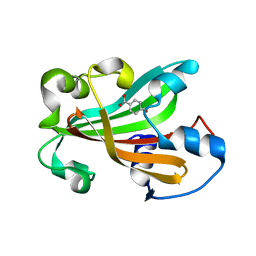 | | CHORISMATE LYASE (WILD-TYPE) WITH BOUND PRODUCT | | Descriptor: | CHORISMATE LYASE, P-HYDROXYBENZOIC ACID | | Authors: | Gallagher, D.T, Mayhew, M, Holden, M.J, Kim, K.J, Howard, A, Vilker, V.L. | | Deposit date: | 2000-10-11 | | Release date: | 2001-04-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of chorismate lyase shows a new fold and a tightly retained product.
Proteins, 44, 2001
|
|
1IXA
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF THE FIRST EGF-LIKE MODULE OF HUMAN FACTOR IX: COMPARISON WITH EGF AND TGF-A | | Descriptor: | EGF-LIKE MODULE OF HUMAN FACTOR IX | | Authors: | Baron, M, Norman, D.G, Harvey, T.S, Hanford, P.A, Mayhew, M, Tse, A.G.D, Brownlee, G.G, Campbell, I.D.C. | | Deposit date: | 1991-11-14 | | Release date: | 1993-10-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the first EGF-like module of human factor IX: comparison with EGF and TGF-alpha.
Protein Sci., 1, 1992
|
|
