4PSD
 
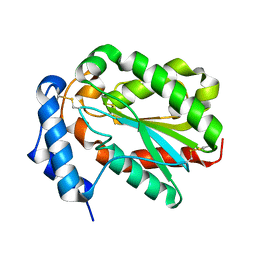 | | Structure of Trichoderma reesei cutinase native form. | | Descriptor: | Carbohydrate esterase family 5 | | Authors: | Roussel, A, Amara, S, Nyyssola, A, Mateos-Diaz, E, Blangy, S, Kontkanen, H, Westerholm-Parvinen, A, Carriere, F, Cambillau, C. | | Deposit date: | 2014-03-07 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Cutinase from Trichoderma reesei with a Lid-Covered Active Site and Kinetic Properties of True Lipases.
J.Mol.Biol., 426, 2014
|
|
4EQQ
 
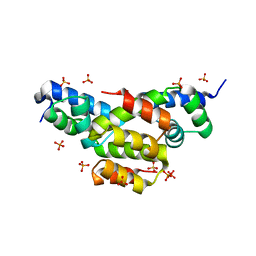 | | Structure of Ltp, a superinfection exclusion protein from the Streptococcus thermophilus temperate phage TP-J34 | | Descriptor: | PHOSPHATE ION, Putative host cell surface-exposed lipoprotein, SULFATE ION | | Authors: | Bebeacua, C, Lorenzo, C, Blangy, S, Spinelli, S, Heller, K, Cambillau, C. | | Deposit date: | 2012-04-19 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray structure of a superinfection exclusion lipoprotein from phage TP-J34 and identification of the tape measure protein as its target.
Mol.Microbiol., 89, 2013
|
|
1TR7
 
 | | FimH adhesin receptor binding domain from uropathogenic E. coli | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, FimH protein, ... | | Authors: | Bouckaert, J, Berglund, J, Schembri, M, De Genst, E, Cools, L, Wuhrer, M, Hung, C.S, Pinkner, J, Slattegard, R, Zavialov, A, Choudhury, D, Langermann, S, Hultgren, S.J, Wyns, L, Klemm, P, Oscarson, S, Knight, S.D, De Greve, H. | | Deposit date: | 2004-06-21 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Receptor binding studies disclose a novel class of high-affinity inhibitors of the Escherichia coli FimH adhesin
Mol.Microbiol., 55, 2005
|
|
3U6X
 
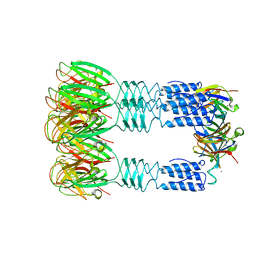 | | Phage TP901-1 baseplate tripod | | Descriptor: | BPP, BROMIDE ION, ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V.I, van Sinderen, D, Cambillau, C. | | Deposit date: | 2011-10-13 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2XC8
 
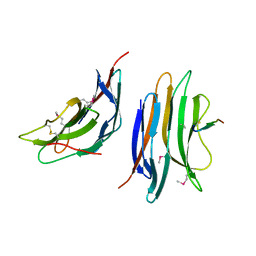 | | Crystal structure of the gene 22 product of the Bacillus subtilis SPP1 phage | | Descriptor: | GENE 22 PRODUCT | | Authors: | Veesler, D, Blangy, S, Tavares, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-04-19 | | Release date: | 2010-06-09 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Spp1 Phage Gp22 Shares Fold Similarity with a Domain of Lactococcal Phage P2 Rbp.
Protein Sci., 19, 2010
|
|
2ACO
 
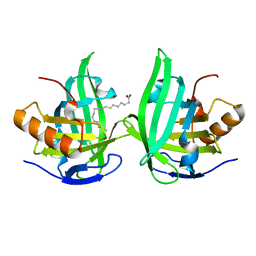 | | Xray structure of Blc dimer in complex with vaccenic acid | | Descriptor: | Outer membrane lipoprotein blc, VACCENIC ACID | | Authors: | Campanacci, V, Bishop, R.E, Reese, L, Blangy, S, Tegoni, M, Cambillau, C. | | Deposit date: | 2005-07-19 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The membrane bound bacterial lipocalin Blc is a functional dimer with binding preference for lysophospholipids.
Febs Lett., 580, 2006
|
|
3D9B
 
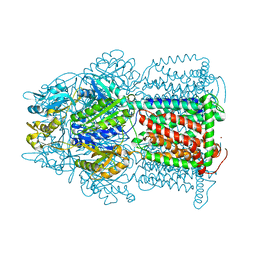 | | Symmetric structure of E. coli AcrB | | Descriptor: | Acriflavine resistance protein B, NICKEL (II) ION | | Authors: | Veesler, D, Blangy, S, Cambillau, C, Sciara, G. | | Deposit date: | 2008-05-27 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | There is a baby in the bath water: AcrB contamination is a major problem in membrane-protein crystallization.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
3RX9
 
 | | 3D structure of SciN from an Escherichia coli Patotype | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Felisberto-Rodrigues, C, Durand, E, Aschtgen, M.-S, Blangy, S, Ortiz-Lombardia, M, Douzy, B, Cambillau, C, Cascales, E. | | Deposit date: | 2011-05-10 | | Release date: | 2011-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Towards a Structural Comprehension of Bacterial Type VI Secretion Systems: Characterization of the TssJ-TssM Complex of an Escherichia coli Pathovar.
Plos Pathog., 7, 2011
|
|
2XF6
 
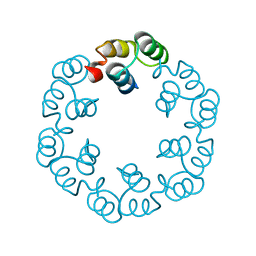 | | Crystal structure of Bacillus subtilis SPP1 phage gp23.1, a putative chaperone. | | Descriptor: | GP23.1 | | Authors: | Veesler, D, Blangy, S, Lichiere, J, Ortiz-Lombardia, M, Tavares, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-05-20 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Spp1 Phage Gp23.1, A Putative Chaperone.
Protein Sci., 19, 2010
|
|
2XF7
 
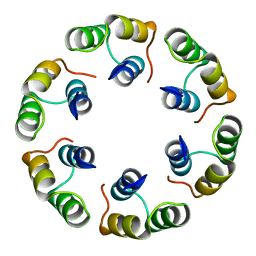 | | Crystal structure of Bacillus subtilis SPP1 phage gp23.1, a putative chaperone. High-resolution structure. | | Descriptor: | GP23.1 | | Authors: | Veesler, D, Blangy, S, Lichiere, J, Ortiz-Lombardia, M, Tavares, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-05-20 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Spp1 Phage Gp23.1, A Putative Chaperone.
Protein Sci., 19, 2010
|
|
2XF5
 
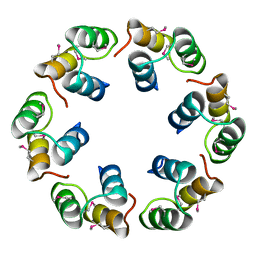 | | Crystal structure of Bacillus subtilis SPP1 phage gp23.1, a putative chaperone. | | Descriptor: | GP23.1 | | Authors: | Veesler, D, Blangy, S, Lichiere, J, Ortiz-Lombardia, M, Tavares, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-05-20 | | Release date: | 2010-08-11 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Spp1 Phage Gp23.1, A Putative Chaperone.
Protein Sci., 19, 2010
|
|
8QUB
 
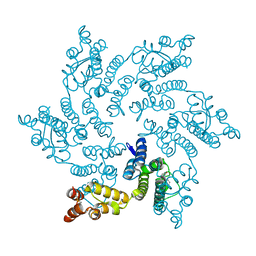 | | Hexameric HIV-1 CA in complex with DDD00074110 | | Descriptor: | (1~{S})-1-phenyl-2,4-dihydro-1~{H}-isoquinolin-3-one, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QUH
 
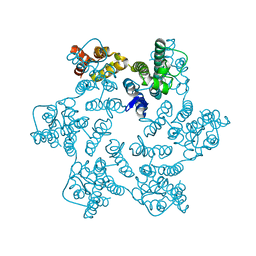 | | Hexameric HIV-1 CA in complex with DDD00057456 | | Descriptor: | 4-methylquinolin-2-ol, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QUJ
 
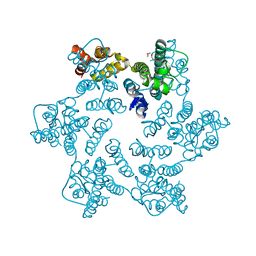 | | Hexameric HIV-1 CA in complex with DDD00100452 | | Descriptor: | 1,2-ETHANEDIOL, 3-(phenylmethyl)-1~{H}-imidazol-2-one, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QUI
 
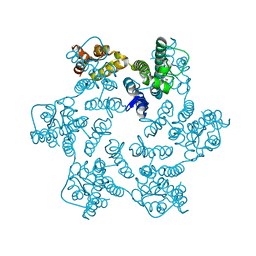 | | Hexameric HIV-1 CA in complex with DDD00024969 | | Descriptor: | Spacer peptide 1, ethyl (3-oxo-2,3-dihydro-4H-1,4-benzoxazin-4-yl)acetate | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QUL
 
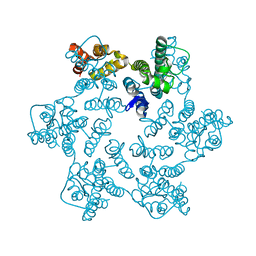 | | Hexameric HIV-1 CA in complex with DDD00100555 | | Descriptor: | 3-(BENZYLOXY)PYRIDIN-2-AMINE, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QUW
 
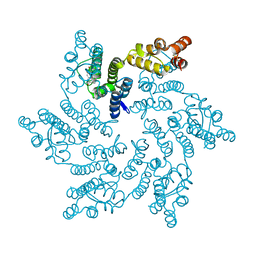 | | Hexameric HIV-1 CA in complex with DDD01044153 | | Descriptor: | (4~{R})-7-oxidanyl-4-phenyl-3,4-dihydro-1~{H}-quinolin-2-one, 1,2-ETHANEDIOL, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QUX
 
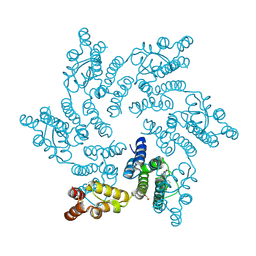 | | Hexameric HIV-1 CA in complex with DDD00100333 | | Descriptor: | 1,2-ETHANEDIOL, 4-benzyl-3,4-dihydroquinoxalin-2(1H)-one, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QV1
 
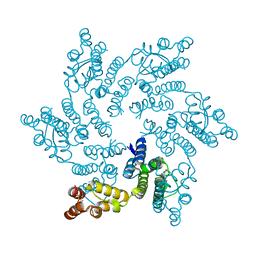 | | Hexameric HIV-1 CA in complex with DDD01728505 | | Descriptor: | Spacer peptide 1, methyl 2-(2-oxidanylidene-1~{H}-quinolin-4-yl)ethanoate | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QV9
 
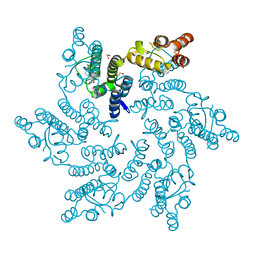 | | Hexameric HIV-1 CA in complex with DDD01829021 | | Descriptor: | 1,2-ETHANEDIOL, 7-bromanyl-3-(phenylmethyl)-1~{H}-benzimidazol-2-one, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QUK
 
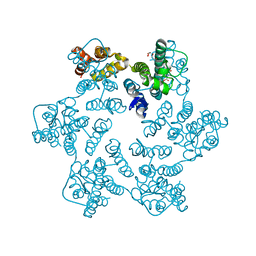 | | Hexameric HIV-1 CA in complex with DDD00100439 | | Descriptor: | (phenylmethyl) 3-oxidanylidenepiperazine-1-carboxylate, 1,2-ETHANEDIOL, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QV4
 
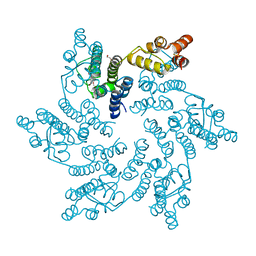 | | Hexameric HIV-1 CA in complex with DDD01728503 | | Descriptor: | 1,2-ETHANEDIOL, Spacer peptide 1, ethyl 2-(3-oxidanylidene-2,4-dihydroquinoxalin-1-yl)ethanoate | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QUY
 
 | | Hexameric HIV-1 CA in complex with DDD01728501 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-methylphenyl)methyl]-1~{H}-quinoxaline-2,3-dione, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
8QVA
 
 | | Hexameric HIV-1 CA in complex with DDD01829894 | | Descriptor: | 1,2-ETHANEDIOL, 7-azanyl-3-(phenylmethyl)-1~{H}-benzimidazol-2-one, Spacer peptide 1 | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Application of an NMR/Crystallography Fragment Screening Platform for the Assessment and Rapid Discovery of New HIV-CA Binding Fragments.
Chemmedchem, 19, 2024
|
|
5W42
 
 | |
