7XWF
 
 | | RLGSGG-AtPRT6 UBR box (highest resolution) | | Descriptor: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | Authors: | Kim, L, Song, H.K. | | Deposit date: | 2022-05-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7Y70
 
 | | RLGSGG-AtPRT6 UBR box (P4332) | | Descriptor: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | Authors: | Kim, L, Song, H.K. | | Deposit date: | 2022-06-21 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7XWD
 
 | | Apo-AtPRT6 UBR box | | Descriptor: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | Authors: | Kim, L, Song, H.K. | | Deposit date: | 2022-05-26 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7Y6Y
 
 | | RLGSGG-AtPRT6 UBR box (C121) | | Descriptor: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | Authors: | Kim, L, Song, H.K. | | Deposit date: | 2022-06-21 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7Y6X
 
 | | RRGSGG-AtPRT6 UBR box (P32) | | Descriptor: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | Authors: | Kim, L, Song, H.K. | | Deposit date: | 2022-06-21 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7Y6W
 
 | | RRGSGG-AtPRT6 UBR box (I222) | | Descriptor: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | Authors: | Kim, L, Song, H.K. | | Deposit date: | 2022-06-21 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
8JBQ
 
 | |
5CA7
 
 | |
5CB1
 
 | | Apo enzyme of human Polymerase lambda | | Descriptor: | DNA polymerase lambda | | Authors: | Liu, M.S, Tsai, M.D. | | Deposit date: | 2015-06-30 | | Release date: | 2016-02-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Mechanism for the Fidelity Modulation of DNA Polymerase lambda
J.Am.Chem.Soc., 138, 2016
|
|
4XRH
 
 | |
4XQ8
 
 | |
7CN9
 
 | | Cryo-EM structure of SARS-CoV-2 Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ho, M, Chang, Y, Wang, C, Wu, Y, Huang, H, Chen, T, Lo, J.M, Chen, X, Ma, C. | | Deposit date: | 2020-07-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | A Carbohydrate-Binding Protein from the Edible Lablab Beans Effectively Blocks the Infections of Influenza Viruses and SARS-CoV-2.
Cell Rep, 32, 2020
|
|
8IO9
 
 | |
8IOA
 
 | |
8IO8
 
 | |
8IO7
 
 | |
8IO6
 
 | |
8IOE
 
 | |
9D93
 
 | |
9D9X
 
 | |
9D94
 
 | |
9D9L
 
 | |
9D9W
 
 | |
7XGZ
 
 | | Cryo-EM structure of the T=4 lake sinai virus 2 virus-like capsid at pH 7.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-04-07 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions
Nat Commun, 14, 2023
|
|
7XPE
 
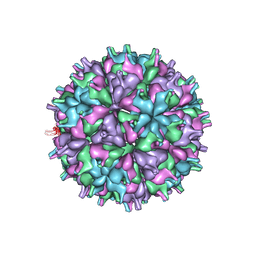 | | Cryo-EM structure of the T=4 lake sinai virus 2 virus-like capsid at pH 8.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-05-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions.
Nat Commun, 14, 2023
|
|
