5IY7
 
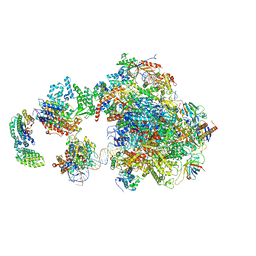 | | Human holo-PIC in the open state | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | Deposit date: | 2016-03-24 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
2JT4
 
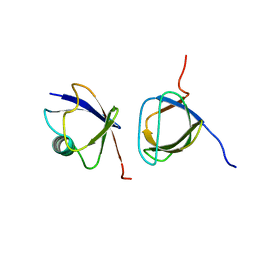 | |
2JU8
 
 | | Solution-State Structures of Oleate-Liganded LFABP, Major Form of 1:2 Protein-Ligand Complex | | Descriptor: | Fatty acid-binding protein, liver, OLEIC ACID | | Authors: | He, Y, Yang, X, Wang, H, Estephan, R, Francis, F, Kodukula, S, Storch, J, Stark, R.E. | | Deposit date: | 2007-08-15 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution-State Molecular Structure of Apo and Oleate-Liganded Liver Fatty Acid-Binding Protein
Biochemistry, 46, 2007
|
|
2KDP
 
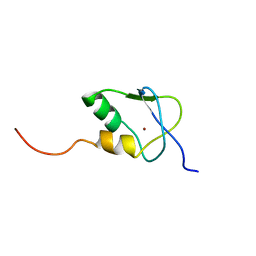 | | Solution Structure of the SAP30 zinc finger motif | | Descriptor: | Histone deacetylase complex subunit SAP30, ZINC ION | | Authors: | He, Y, Imhoff, R, Sahu, A, Radhakrishnan, I. | | Deposit date: | 2009-01-14 | | Release date: | 2009-03-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel zinc finger motif in the SAP30 polypeptide of the Sin3 corepressor complex and its potential role in nucleic acid recognition
Nucleic Acids Res., 37, 2009
|
|
2KY4
 
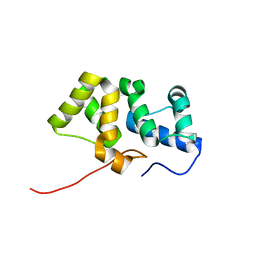 | | Solution NMR structure of the PBS linker domain of phycobilisome linker polypeptide from Anabaena sp. Northeast Structural Genomics Consortium Target NsR123E | | Descriptor: | Phycobilisome linker polypeptide | | Authors: | He, Y, Eletsky, A, Mills, J.L, Lee, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Lee, H, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-05-14 | | Release date: | 2010-07-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the PBS linker domain of phycobilisome linker polypeptide from Anabaena sp. Northeast Structural Genomics Consortium Target NsR123E
To be Published
|
|
2KRU
 
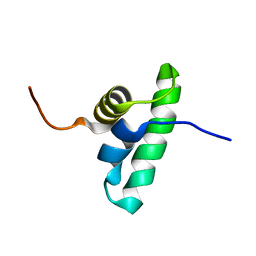 | | Solution NMR structure of the PCP_red domain of light-independent protochlorophyllide reductase subunit B from Chlorobium tepidum. Northeast Structural Genomics Consortium Target CtR69A | | Descriptor: | Light-independent protochlorophyllide reductase subunit B | | Authors: | He, Y, Eletsky, A, Lee, D, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-12-22 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the PCP_red domain of light-independent protochlorophyllide reductase subunit B from Chlorobium tepidum. Northeast Structural Genomics Consortium Target CtR69A
To be Published
|
|
2KYW
 
 | | Solution NMR Structure of a domain of adhesion exoprotein from Pediococcus pentosaceus, Northeast Structural Genomics Consortium Target PtR41O | | Descriptor: | Adhesion exoprotein | | Authors: | He, Y, Eletsky, A, Mills, J.L, Wang, H, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Lee, H.-W, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of a domain of adhesion exoprotein from Pediococcus pentosaceus, Northeast Structural Genomics Consortium Target PtR41O
To be Published
|
|
7UY6
 
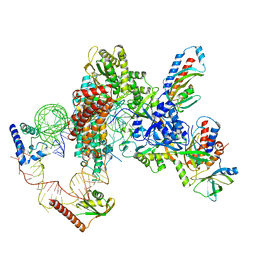 | | Tetrahymena telomerase at 2.9 Angstrom resolution | | Descriptor: | Telomerase La-related protein p65, Telomerase RNA, Telomerase associated protein p50, ... | | Authors: | He, Y, Song, H, Chan, H, Wang, Y, Liu, B, Susac, L, Zhou, Z.H, Feigon, J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-07-13 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of Tetrahymena telomerase-bound CST with polymerase alpha-primase.
Nature, 608, 2022
|
|
7UY7
 
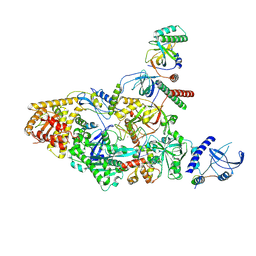 | | Tetrahymena CST with Polymerase alpha-Primase | | Descriptor: | DNA polymerase, Telomerase associated protein p50, Telomerase-associated protein of 19 kDa, ... | | Authors: | He, Y, Song, H, Chan, H, Wang, Y, Liu, B, Susac, L, Zhou, Z.H, Feigon, J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-07-13 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of Tetrahymena telomerase-bound CST with polymerase alpha-primase.
Nature, 608, 2022
|
|
7UY5
 
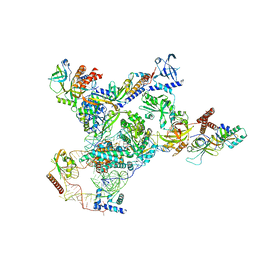 | | Tetrahymena telomerase with CST | | Descriptor: | Telomerase La-related protein p65, Telomerase RNA, Telomerase associated protein p50, ... | | Authors: | He, Y, Song, H, Chan, H, Wang, Y, Liu, B, Susac, L, Zhou, Z.H, Feigon, J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-07-13 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of Tetrahymena telomerase-bound CST with polymerase alpha-primase.
Nature, 608, 2022
|
|
7UY8
 
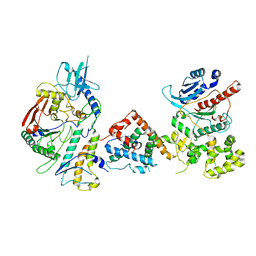 | | Tetrahymena Polymerase alpha-Primase | | Descriptor: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | He, Y, Song, H, Chan, H, Wang, Y, Liu, B, Susac, L, Zhou, Z.H, Feigon, J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-07-13 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of Tetrahymena telomerase-bound CST with polymerase alpha-primase.
Nature, 608, 2022
|
|
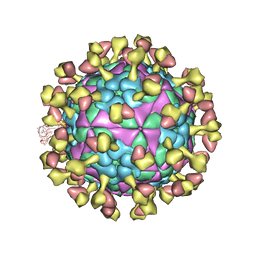
7D3M
 
 | |
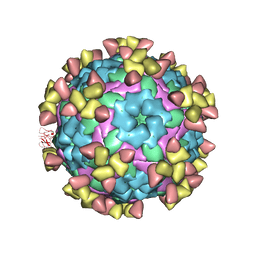
7D3K
 
 | |
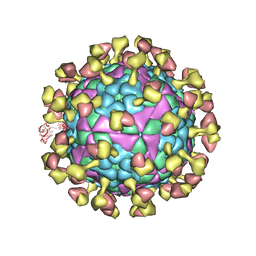
7D3R
 
 | |
7D3L
 
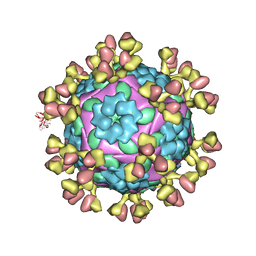 | |
7FEJ
 
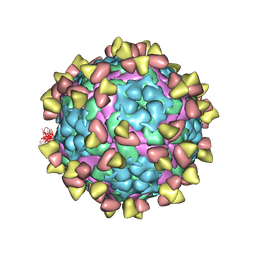 | | Complex of FMDV A/AF/72 and bovine neutralizing scFv antibody R55 | | Descriptor: | A/AF/72 VP1, A/AF/72 VP2, A/AF/72 VP3, ... | | Authors: | He, Y, Li, K, Lou, Z. | | Deposit date: | 2021-07-20 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structures of Foot-and-Mouth Disease Virus with Bovine Neutralizing Antibodies Reveal the Determinant of Intraserotype Cross-Neutralization.
J.Virol., 95, 2021
|
|
7FEI
 
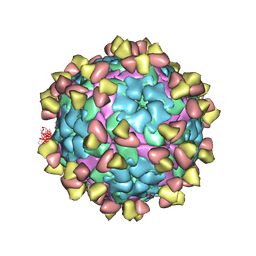 | | Complex of FMDV A/WH/CHA/09 and bovine neutralizing scFv antibody R55 | | Descriptor: | Capsid protein VP0, IG HEAVY CHAIN VARIABLE REGION, IG LAMDA CHAIN VARIABLE REGION | | Authors: | He, Y, Li, K, Lou, Z. | | Deposit date: | 2021-07-20 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structures of Foot-and-Mouth Disease Virus with Bovine Neutralizing Antibodies Reveal the Determinant of Intraserotype Cross-Neutralization.
J.Virol., 95, 2021
|
|
8K0Y
 
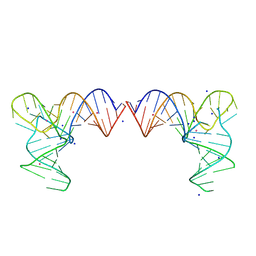 | |
8K2Z
 
 | |
8K30
 
 | |
8K1E
 
 | | Crystal structure of a human menRNA | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (58-MER), ... | | Authors: | He, Y, Deng, J, Lin, X, Huang, L. | | Deposit date: | 2023-07-11 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of mascRNA and menRNA
To Be Published
|
|
7MP7
 
 | | Rules for designing protein fold switches and their implications for the folding code | | Descriptor: | Sb3 | | Authors: | He, Y, Chen, Y, Ruan, B, Choi, J, Chen, Y, Motabar, D, Solomon, T, Simmerman, R, Kauffman, T, Gallagher, T, Bryan, P, Orban, J. | | Deposit date: | 2021-05-04 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of a protein fold switching network.
Nat Commun, 14, 2023
|
|
7MN1
 
 | | Rules for designing protein fold switches and their implications for the folding code | | Descriptor: | Sa1 | | Authors: | He, Y, Chen, Y, Ruan, B, Choi, J, Chen, Y, Motabar, D, Solomon, T, Simmerman, R, Kauffman, T, Gallagher, T, Bryan, P, Orban, J. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of a protein fold switching network.
Nat Commun, 14, 2023
|
|
7MQ4
 
 | | Rules for designing protein fold switches and their implications for the folding code | | Descriptor: | Sb1 | | Authors: | He, Y, Chen, Y, Ruan, B, Choi, J, Chen, Y, Motabar, D, Solomon, T, Simmerman, R, Kauffman, T, Gallagher, T, Bryan, P, Orban, J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of a protein fold switching network.
Nat Commun, 14, 2023
|
|
9LJJ
 
 | | cryo-EM structure of a nanobody bound heliorhodopsin | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, ACETATE ION, Heliorhodopsin, ... | | Authors: | He, Y, Xia, R. | | Deposit date: | 2025-01-15 | | Release date: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Cryo-EM structure of a nanobody-bound heliorhodopsin.
Biochem.Biophys.Res.Commun., 750, 2025
|
|
