1YIZ
 
 | | Aedes aegypti kynurenine aminotrasferase | | Descriptor: | BROMIDE ION, kynurenine aminotransferase; glutamine transaminase | | Authors: | Han, Q, Gao, Y.G, Robinson, H, Ding, H, Wilson, S, Li, J. | | Deposit date: | 2005-01-13 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of Aedes aegypti kynurenine aminotransferase.
FEBS J., 272, 2005
|
|
7WI4
 
 | | Cryo-EM structure of E.Coli FtsH protease cytosolic domains | | Descriptor: | ATP-dependent zinc metalloprotease FtsH, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Qiao, Z, Gao, Y.G. | | Deposit date: | 2022-01-02 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the entire FtsH-HflKC AAA protease complex.
Cell Rep, 39, 2022
|
|
7WI3
 
 | | Cryo-EM structure of E.Coli FtsH-HflkC AAA protease complex | | Descriptor: | ATP-dependent zinc metalloprotease FtsH, Modulator of FtsH protease HflC, Modulator of FtsH protease HflK | | Authors: | Qiao, Z, Gao, Y.G. | | Deposit date: | 2022-01-02 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the entire FtsH-HflKC AAA protease complex.
Cell Rep, 39, 2022
|
|
5IMR
 
 | | Structure of ribosome bound to cofactor at 5.7 angstrom resolution | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, V, Ero, R, Jian, G.K, Ahmed, T, Zhan, Y, Bhushan, S, Gao, Y.G. | | Deposit date: | 2016-03-06 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structure of the GTP Form of Elongation Factor 4 (EF4) Bound to the Ribosome
J.Biol.Chem., 291, 2016
|
|
5IMQ
 
 | | Structure of ribosome bound to cofactor at 3.8 angstrom resolution | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, V, Ero, R, Jian, G.K, Ahmed, T, Zhan, Y, Bhushan, S, Gao, Y.G. | | Deposit date: | 2016-03-06 | | Release date: | 2016-05-18 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the GTP Form of Elongation Factor 4 (EF4) Bound to the Ribosome
J.Biol.Chem., 291, 2016
|
|
6KFS
 
 | | Structure of CCM related protein | | Descriptor: | LCIB_C_CA domain-containing protein, ZINC ION | | Authors: | Jin, S, Gao, Y.G. | | Deposit date: | 2019-07-08 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and biochemical characterization of novel carbonic anhydrases from Phaeodactylum tricornutum.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6IUY
 
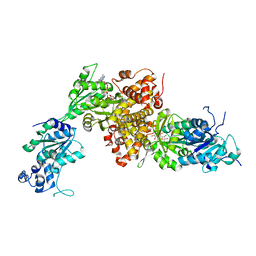 | | Structure of DsGPDH of Dunaliella salina | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, GLYCEROL, Glycerol-3-phosphate dehydrogenase [NAD(+)], ... | | Authors: | He, Q, Toh, J.D, Ero, R, Qiao, Z, Kumar, V, Gao, Y.G. | | Deposit date: | 2018-12-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The unusual di-domain structure of Dunaliella salina glycerol-3-phosphate dehydrogenase enables direct conversion of dihydroxyacetone phosphate to glycerol.
Plant J., 102, 2020
|
|
6IEJ
 
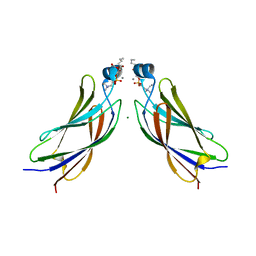 | | The C2 domain of cytosolic phospholipase A2 alpha bound to phosphatidylcholine | | Descriptor: | 1,2-dihexanoyl-sn-glycero-3-phosphocholine, CALCIUM ION, Cytosolic phospholipase A2, ... | | Authors: | Hirano, Y, Gao, Y.G, Stephenson, D.J, Vu, N.T, Malinina, L, Chalfant, C.E, Patel, D.J, Brown, R.E. | | Deposit date: | 2018-09-14 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structural basis of phosphatidylcholine recognition by the C2-domain of cytosolic phospholipase A2alpha.
Elife, 8, 2019
|
|
7VBW
 
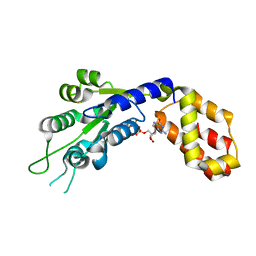 | |
7VBS
 
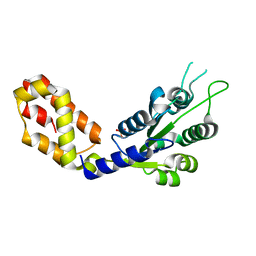 | |
7WQ5
 
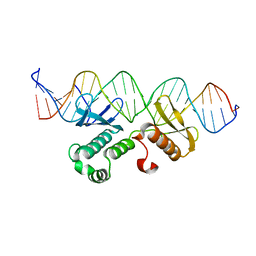 | | Crystal structure of Arabidopsis transcriptional factor WRINKLED1 with dsDNA | | Descriptor: | AMMONIUM ION, DNA (5'-D(P*GP*TP*GP*GP*AP*CP*GP*AP*TP*GP*AP*AP*AP*CP*CP*GP*AP*GP*GP*AP*AP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*TP*TP*CP*CP*TP*CP*GP*GP*TP*TP*TP*CP*AP*TP*CP*GP*TP*CP*CP*AP*C)-3'), ... | | Authors: | Zhu, Q, Gao, Y.G. | | Deposit date: | 2022-01-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis of the key regulator WRINKLED1 in plant oil biosynthesis.
Sci Adv, 8, 2022
|
|
7C3M
 
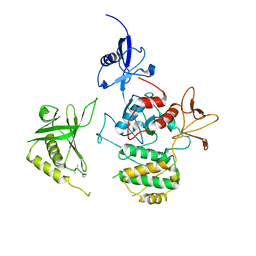 | | Structure of FERM protein | | Descriptor: | Fermitin family homolog 3,Fermitin family homolog 3,Fermitin family homolog 3 | | Authors: | Bu, W, Loh, Z.Y, Jin, S, Basu, S, Ero, R, Park, J.E, Yan, X, Wang, M, Sze, S.K, Tan, S.M, Gao, Y.G. | | Deposit date: | 2020-05-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis of human full-length kindlin-3 homotrimer in an auto-inhibited state.
Plos Biol., 18, 2020
|
|
5Y4R
 
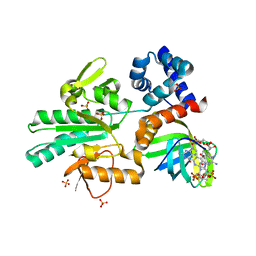 | | Structure of a methyltransferase complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Chemotaxis protein methyltransferase 1, Cyclic diguanosine monophosphate-binding protein PA4608, ... | | Authors: | Yan, X, Xin, L, Tan, Y.J, Jin, S, Liang, Z.X, Gao, Y.G. | | Deposit date: | 2017-08-04 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Structural analyses unravel the molecular mechanism of cyclic di-GMP regulation of bacterial chemotaxis via a PilZ adaptor protein.
J. Biol. Chem., 293, 2018
|
|
6ABR
 
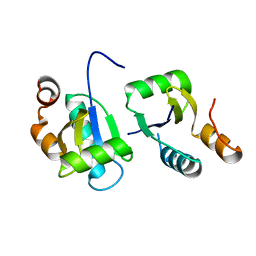 | | Actin interacting protein 5 (Aip5, wild type) | | Descriptor: | Actin binding protein | | Authors: | Sun, J, Xie, Y, Toh, J.D.W, Hong, W, MIao, Y, Gao, Y.G. | | Deposit date: | 2018-07-23 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Polarisome scaffolder Spa2-mediated macromolecular condensation of Aip5 for actin polymerization.
Nat Commun, 10, 2019
|
|
7CK1
 
 | | Crystal structure of arabidopsis CESA3 catalytic domain | | Descriptor: | Cellulose synthase A catalytic subunit 3 [UDP-forming],Cellulose synthase A catalytic subunit 3 [UDP-forming], MANGANESE (II) ION | | Authors: | Qiao, Z, Gao, Y.G. | | Deposit date: | 2020-07-15 | | Release date: | 2021-03-17 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Arabidopsis CESA3 catalytic domain with its substrate UDP-glucose provides insight into the mechanism of cellulose synthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CK3
 
 | | Crystal structure of Arabidopsis CESA3 catalytic domain | | Descriptor: | Cellulose synthase A catalytic subunit 3 [UDP-forming],Cellulose synthase A catalytic subunit 3 [UDP-forming] | | Authors: | Qiao, Z, Gao, Y.G. | | Deposit date: | 2020-07-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of Arabidopsis CESA3 catalytic domain with its substrate UDP-glucose provides insight into the mechanism of cellulose synthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CK2
 
 | | Crystal structure of Arabidopsis CESA3 catalytic domain with UDP-Glucose | | Descriptor: | Cellulose synthase A catalytic subunit 3 [UDP-forming],Cellulose synthase A catalytic subunit 3 [UDP-forming], MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Qiao, Z, Gao, Y.G. | | Deposit date: | 2020-07-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Arabidopsis CESA3 catalytic domain with its substrate UDP-glucose provides insight into the mechanism of cellulose synthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6ABS
 
 | | Actin interacting protein 5 (Aip5, mutant) | | Descriptor: | Actin binding protein, PENTAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | Sun, J, Ying, X, Toh, J, Hong, W, Miao, Y, Gao, Y.G. | | Deposit date: | 2018-07-23 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Polarisome scaffolder Spa2-mediated macromolecular condensation of Aip5 for actin polymerization.
Nat Commun, 10, 2019
|
|
7CE1
 
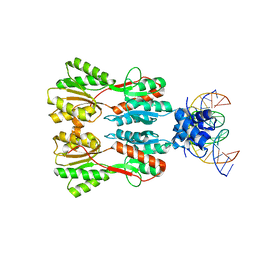 | | Complex STRUCTURE OF TRANSCRIPTION FACTOR SghR with its COGNATE DNA | | Descriptor: | LacI-type transcription factor, promoter DNA | | Authors: | Ye, F.Z, Wang, C, Yan, X.F, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2020-06-21 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of a novel repressor, SghR, controllingAgrobacteriuminfection by cross-talking to plants.
J.Biol.Chem., 295, 2020
|
|
5Y4S
 
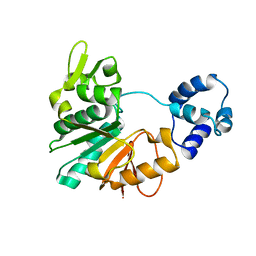 | | Structure of a methyltransferase complex | | Descriptor: | Chemotaxis protein methyltransferase 1 | | Authors: | Yan, X, Xin, L, Tan, Y.J, Jin, S, Liang, Z.X, Gao, Y.G. | | Deposit date: | 2017-08-04 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.405 Å) | | Cite: | Structural analyses unravel the molecular mechanism of cyclic di-GMP regulation of bacterial chemotaxis via a PilZ adaptor protein.
J. Biol. Chem., 293, 2018
|
|
7CDV
 
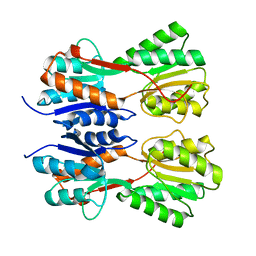 | | STRUCTURE OF A NOVEL VIRULENCE REGULATION FACTOR SghR | | Descriptor: | LacI-type transcription factor | | Authors: | Ye, F.Z, Wang, C, Yan, X.F, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of a novel repressor, SghR, controllingAgrobacteriuminfection by cross-talking to plants.
J.Biol.Chem., 295, 2020
|
|
7CDX
 
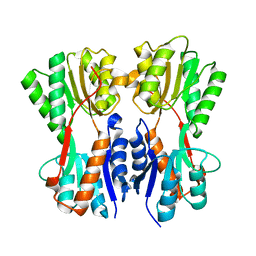 | | Complex STRUCTURE OF A NOVEL VIRULENCE REGULATION FACTOR SghR with its effector sucrose | | Descriptor: | LacI-type transcription factor, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Ye, F.Z, Wang, C, Yan, X.F, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Structural basis of a novel repressor, SghR, controllingAgrobacteriuminfection by cross-talking to plants.
J.Biol.Chem., 295, 2020
|
|
215D
 
 | |
236D
 
 | |
277D
 
 | |
