6N2C
 
 | |
6N6P
 
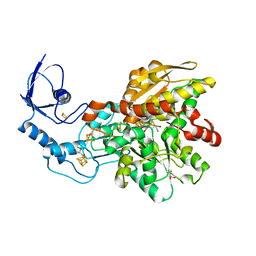 | | Crystal structure of [FeFe]-hydrogenase in the presence of 7 mM Sodium dithionite | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Zadvornyy, O.A, Keable, S.M, Peters, J.W. | | Deposit date: | 2018-11-26 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tuning Catalytic Bias of Hydrogen Gas Producing Hydrogenases.
J.Am.Chem.Soc., 142, 2020
|
|
6NAC
 
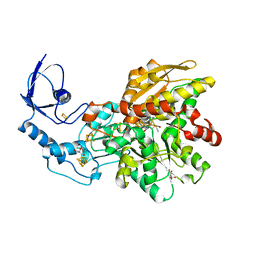 | | Crystal structure of [FeFe]-hydrogenase I (CpI) solved with single pulse free electron laser data | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Cohen, A.E, Davidson, C.M, Zadvornyy, O.A, Keable, S.M, Lyubimov, A.Y, Song, J, McPhillips, S.E, Soltis, S.M, Peters, J.W. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Tuning Catalytic Bias of Hydrogen Gas Producing Hydrogenases.
J.Am.Chem.Soc., 142, 2020
|
|
6D5D
 
 | |
6N2B
 
 | |
6N59
 
 | | 1.0 Angstrom crystal structure of [FeFe]-hydrogenase | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Zadvornyy, O.A, Keable, S.M, Artz, J.H, Peters, J.W. | | Deposit date: | 2018-11-21 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Tuning Catalytic Bias of Hydrogen Gas Producing Hydrogenases.
J.Am.Chem.Soc., 142, 2020
|
|
6CFW
 
 | |
1BVU
 
 | | GLUTAMATE DEHYDROGENASE FROM THERMOCOCCUS LITORALIS | | Descriptor: | PROTEIN (GLUTAMATE DEHYDROGENASE) | | Authors: | Baker, P.J, Britton, K.L, Yip, K.S, Stillman, T.J, Rice, D.W. | | Deposit date: | 1999-07-20 | | Release date: | 1999-09-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure determination of the glutamate dehydrogenase from the hyperthermophile Thermococcus litoralis and its comparison with that from Pyrococcus furiosus
J.Mol.Biol., 293, 1999
|
|
1CAD
 
 | | X-RAY CRYSTAL STRUCTURES OF THE OXIDIZED AND REDUCED FORMS OF THE RUBREDOXIN FROM THE MARINE HYPERTHERMOPHILIC ARCHAEBACTERIUM PYROCOCCUS FURIOSUS | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Day, M.W, Hsu, B.T, Joshua-Tor, L, Park, J.B, Zhou, Z.H, Adams, M.W.W, Rees, D.C. | | Deposit date: | 1992-05-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structures of the oxidized and reduced forms of the rubredoxin from the marine hyperthermophilic archaebacterium Pyrococcus furiosus.
Protein Sci., 1, 1992
|
|
9NEZ
 
 | | Structure of the Pyrococcus furiosus SHI complex | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Xiao, X, Li, H. | | Deposit date: | 2025-02-20 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural insights into the biotechnologically relevant reversible NADPH-oxidizing NiFe-hydrogenase from P.furiosus.
Structure, 2025
|
|
9NF0
 
 | | Structure of the NADPH-bound Pyrococcus furiosus SHI complex | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Xiao, X, Li, H. | | Deposit date: | 2025-02-20 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural insights into the biotechnologically relevant reversible NADPH-oxidizing NiFe-hydrogenase from P.furiosus.
Structure, 2025
|
|
7KOE
 
 | | Electron bifurcating flavoprotein Fix/EtfABCX | | Descriptor: | Electron transfer flavoprotein, alpha subunit, beta subunit, ... | | Authors: | Feng, X, Li, H. | | Deposit date: | 2020-11-08 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryoelectron microscopy structure and mechanism of the membrane-associated electron-bifurcating flavoprotein Fix/EtfABCX.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6X1O
 
 | | WOR5 from Pyrococcus furiosus, as crystallized | | Descriptor: | (1R)-1-hydroxybutane-1-sulfonic acid, CHLORIDE ION, Formaldehyde:ferredoxin oxidoreductase wor5, ... | | Authors: | Mathew, L.G, Lanzilotta, W.N. | | Deposit date: | 2020-05-19 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | An unprecedented function for a tungsten-containing oxidoreductase.
J.Biol.Inorg.Chem., 27, 2022
|
|
6X6U
 
 | | WOR5 from Pyrococcus furiosus, taurine-bound | | Descriptor: | 2-AMINOETHANESULFONIC ACID, CHLORIDE ION, Formaldehyde:ferredoxin oxidoreductase wor5, ... | | Authors: | Lanzilotta, W.N, Mathew, L.G. | | Deposit date: | 2020-05-29 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | An unprecedented function for a tungsten-containing oxidoreductase.
J.Biol.Inorg.Chem., 27, 2022
|
|
7T30
 
 | |
7T2R
 
 | |
9CTD
 
 | | CtfAB F42TS45C mutant co-crystallized with acetyl-CoA | | Descriptor: | 3-oxoacid CoA-transferase, A subunit, B subunit, ... | | Authors: | Buhrman, G, Bing, R. | | Deposit date: | 2024-07-24 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and kinetic characterization of an acetoacetyl-Coenzyme A: acetate Coenzyme A transferase from the extreme thermophile Thermosipho melanesiensis.
Biochem.J., 482, 2025
|
|
9CQ2
 
 | | CtfAB E46D active site mutant hydrolase | | Descriptor: | 3-oxoacid CoA-transferase, A subunit, B subunit, ... | | Authors: | Buhrman, G, Bing, R. | | Deposit date: | 2024-07-19 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and kinetic characterization of an acetoacetyl-Coenzyme A: acetate Coenzyme A transferase from the extreme thermophile Thermosipho melanesiensis.
Biochem.J., 482, 2025
|
|
9CRY
 
 | | CtfAB E46S active site mutant inactive | | Descriptor: | 3-oxoacid CoA-transferase, A subunit, B subunit, ... | | Authors: | Buhrman, G, Bing, R. | | Deposit date: | 2024-07-22 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and kinetic characterization of an acetoacetyl-Coenzyme A: acetate Coenzyme A transferase from the extreme thermophile Thermosipho melanesiensis.
Biochem.J., 482, 2025
|
|
9CSC
 
 | | CtfAB Native Acetoacetate-CoA Transferase protein | | Descriptor: | 3-oxoacid CoA-transferase, A subunit, B subunit, ... | | Authors: | Buhrman, G, Bing, R. | | Deposit date: | 2024-07-23 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and kinetic characterization of an acetoacetyl-Coenzyme A: acetate Coenzyme A transferase from the extreme thermophile Thermosipho melanesiensis.
Biochem.J., 482, 2025
|
|
9D2C
 
 | | Crystal Structure of Tungbindin Treated with Proteinase K | | Descriptor: | MOLYBDATE ION, Molybdenum-pterin binding domain-containing protein | | Authors: | Zhou, D, Chen, L, Rose, J.P, Wang, B.C. | | Deposit date: | 2024-08-08 | | Release date: | 2024-09-04 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Storage of the vital metal tungsten in a dominant SCFA-producing human gut microbe Eubacterium limosum and implications for other gut microbes.
Mbio, 16, 2025
|
|
9BP5
 
 | |
9BOV
 
 | |
3MPS
 
 | | Peroxide Bound Oxidized Rubrerythrin from Pyrococcus furiosus | | Descriptor: | FE (III) ION, HYDROGEN PEROXIDE, MU-OXO-DIIRON, ... | | Authors: | Dillard, B.D, Adams, M.W.W, Lanzilotta, W.N. | | Deposit date: | 2010-04-27 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A cryo-crystallographic time course for peroxide reduction by rubrerythrin from Pyrococcus furiosus.
J.Biol.Inorg.Chem., 16, 2011
|
|
1VK1
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-392566-001 | | Descriptor: | Conserved hypothetical protein, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Shah, A, Liu, Z.J, Tempel, W, Chen, L, Lee, D, Yang, H, Chang, J, Zhao, M, Ng, J, Rose, J, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-04-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | (NZ)CH...O contacts assist crystallization of a ParB-like nuclease.
Bmc Struct.Biol., 7, 2007
|
|
