6B9H
 
 | |
6BCY
 
 | |
6BD1
 
 | |
6C1D
 
 | | High-Resolution Cryo-EM Structures of Actin-bound Myosin States Reveal the Mechanism of Myosin Force Sensing | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mentes, A, Huehn, A, Liu, X, Zwolak, A, Dominguez, R, Shuman, H, Ostap, E.M, Sindelar, C.V. | | Deposit date: | 2018-01-04 | | Release date: | 2018-01-31 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C1G
 
 | | High-Resolution Cryo-EM Structures of Actin-bound Myosin States Reveal the Mechanism of Myosin Force Sensing | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mentes, A, Huehn, A, Liu, X, Zwolak, A, Dominguez, R, Shuman, H, Ostap, E.M, Sindelar, C.V. | | Deposit date: | 2018-01-04 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C1H
 
 | | High-Resolution Cryo-EM Structures of Actin-bound Myosin States Reveal the Mechanism of Myosin Force Sensing | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mentes, A, Huehn, A, Liu, X, Zwolak, A, Dominguez, R, Shuman, H, Ostap, E.M, Sindelar, C.V. | | Deposit date: | 2018-01-04 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BQT
 
 | |
3E5H
 
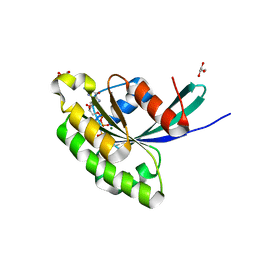 | | Crystal Structure of Rab28 GTPase in the Active (GppNHp-bound) Form | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Lee, S.H, Baek, K, Li, Y, Dominguez, R. | | Deposit date: | 2008-08-13 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Large nucleotide-dependent conformational change in Rab28.
Febs Lett., 582, 2008
|
|
6UHC
 
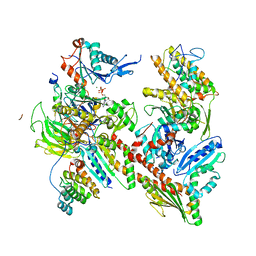 | | CryoEM structure of human Arp2/3 complex with bound NPFs | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1B, ... | | Authors: | Zimmet, A, van Eeuwen, T, Dominguez, R. | | Deposit date: | 2019-09-27 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of NPF-bound human Arp2/3 complex and activation mechanism.
Sci Adv, 6, 2020
|
|
1TN4
 
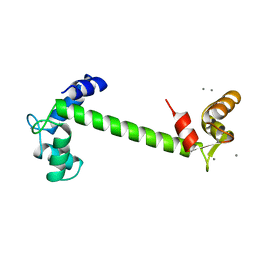 | | FOUR CALCIUM TNC | | Descriptor: | CALCIUM ION, TROPONIN C | | Authors: | Love, M.L, Dominguez, R, Houdusse, A, Cohen, C. | | Deposit date: | 1997-09-18 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of four Ca2+-bound troponin C at 2.0 A resolution: further insights into the Ca2+-switch in the calmodulin superfamily.
Structure, 5, 1997
|
|
1S70
 
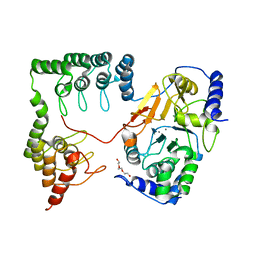 | | Complex between protein ser/thr phosphatase-1 (delta) and the myosin phosphatase targeting subunit 1 (MYPT1) | | Descriptor: | 130 kDa myosin-binding subunit of smooth muscle myosin phophatase (M130), MANGANESE (II) ION, Serine/threonine protein phosphatase PP1-beta (or delta) catalytic subunit, ... | | Authors: | Kerff, F, Terrak, M, Dominguez, R. | | Deposit date: | 2004-01-28 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of protein phosphatase 1 regulation
Nature, 429, 2004
|
|
3T62
 
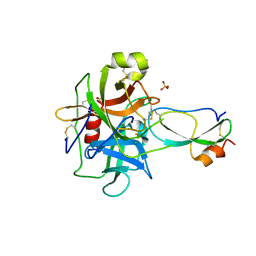 | | Crystal structure of recombinant Kunitz Type serine protease Inhibitor-1 from the Caribbean Sea anemone Stichodactyla helianthus in complex with bovine chymotrypsin | | Descriptor: | Chymotrypsinogen A, Kunitz-type proteinase inhibitor SHPI-1, SULFATE ION | | Authors: | Garcia-Fernandez, R, Dominguez, R, Oberthuer, D, Pons, T, Gonzalez-Gonzalez, Y, Chavez, M.A, Betzel, C, Redecke, L. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into chymotrypsin inhibition by the Kunitz-type inhibitor-1 from the marine invertebrate Stichodactyla helianthus
To be Published
|
|
1M45
 
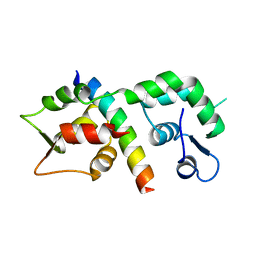 | |
1M46
 
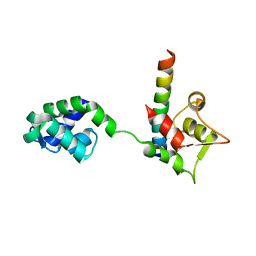 | |
4JS0
 
 | | Complex of Cdc42 with the CRIB-PR domain of IRSp53 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Brain-specific angiogenesis inhibitor 1-associated protein 2, Cell division control protein 42 homolog, ... | | Authors: | Kast, D.J, Dominguez, R. | | Deposit date: | 2013-03-22 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of IRSp53 inhibition and combinatorial activation by Cdc42 and downstream effectors.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4L79
 
 | | Crystal Structure of nucleotide-free Myosin 1b residues 1-728 with bound Calmodulin | | Descriptor: | Calmodulin, MAGNESIUM ION, Unconventional myosin-Ib | | Authors: | Shuman, H, Zwolak, A, Dominguez, R, Ostap, E.M. | | Deposit date: | 2013-06-13 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A vertebrate myosin-I structure reveals unique insights into myosin mechanochemical tuning.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4J7O
 
 | |
4K17
 
 | | Crystal Structure of mouse CARMIL residues 1-668 | | Descriptor: | CHLORIDE ION, GAMMA-AMINO-BUTANOIC ACID, Leucine-rich repeat-containing protein 16A, ... | | Authors: | Zwolak, A, Dominguez, R. | | Deposit date: | 2013-04-04 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.895 Å) | | Cite: | CARMIL leading edge localization depends on a non-canonical PH domain and dimerization.
Nat Commun, 4, 2013
|
|
2D1K
 
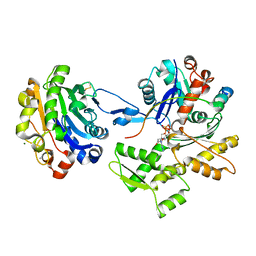 | | Ternary complex of the WH2 domain of mim with actin-dnase I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Chereau, D, Kerff, F, Dominguez, R. | | Deposit date: | 2005-08-26 | | Release date: | 2006-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the actin-binding function of missing-in-metastasis
Structure, 15, 2007
|
|
2D1L
 
 | | Structure of F-actin binding domain IMD of MIM (Missing In Metastasis) | | Descriptor: | Metastasis suppressor protein 1 | | Authors: | Lee, S.H, Kerff, F, Chereau, D, Ferron, F, Dominguez, R. | | Deposit date: | 2005-08-27 | | Release date: | 2006-09-12 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the actin-binding function of missing-in-metastasis
Structure, 15, 2007
|
|
2EYN
 
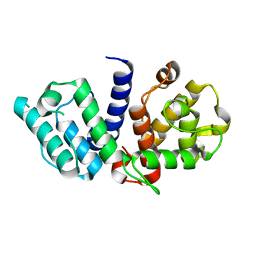 | | Crystal structure of the actin-binding domain of human alpha-actinin 1 at 1.8 Angstrom resolution | | Descriptor: | Alpha-actinin 1 | | Authors: | Borrego-Diaz, E, Kerff, F, Lee, S.H, Ferron, F, Li, Y, Dominguez, R. | | Deposit date: | 2005-11-09 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the actin-binding domain of alpha-actinin 1: Evaluating two competing actin-binding models.
J.Struct.Biol., 155, 2006
|
|
2EYI
 
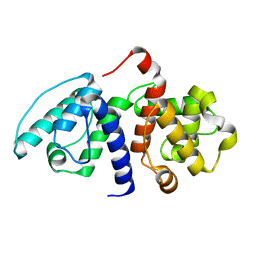 | | Crystal structure of the actin-binding domain of human alpha-actinin 1 at 1.7 Angstrom resolution | | Descriptor: | Alpha-actinin 1 | | Authors: | Borrego-Diaz, E, Kerff, F, Lee, S.H, Ferron, F, Li, Y, Dominguez, R. | | Deposit date: | 2005-11-09 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the actin-binding domain of alpha-actinin 1: Evaluating two competing actin-binding models.
J.Struct.Biol., 155, 2006
|
|
4Z8G
 
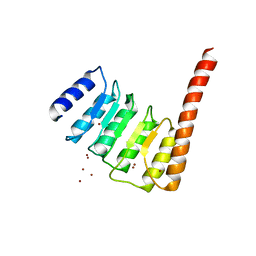 | |
4Z79
 
 | | Leiomodin-1 Actin-Binding Site 2 (ABS2) | | Descriptor: | CHLORIDE ION, GLYCEROL, Leiomodin-1 Actin-Binding Site 2 (ABS2), ... | | Authors: | Rebowski, G, Boczkowska, M, Dominguez, R. | | Deposit date: | 2015-04-06 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | How Leiomodin and Tropomodulin use a common fold for different actin assembly functions.
Nat Commun, 6, 2015
|
|
4Z94
 
 | | Actin Complex With a Chimera of Tropomodulin-1 and Leiomodin-1 Actin-Binding Site 2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rebowski, G, Boczkowska, M, Dominguez, R. | | Deposit date: | 2015-04-09 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | How Leiomodin and Tropomodulin use a common fold for different actin assembly functions.
Nat Commun, 6, 2015
|
|
